FRET enabled real time detection of RNA-small molecule binding
- PMID: 19908830
- PMCID: PMC3031783
- DOI: 10.1021/ja905767g
FRET enabled real time detection of RNA-small molecule binding
Abstract
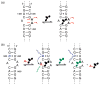
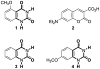
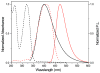
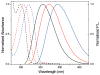
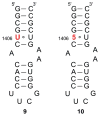
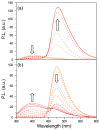
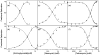
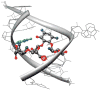
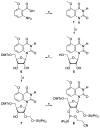
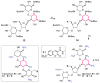
A robust analysis and discovery platform for antibiotics targeting the bacterial rRNA A-site has been developed by incorporating a new emissive U surrogate into the RNA and labeling the aminoglycosides with an appropriate fluorescence acceptor. Specifically, a 5-methoxyquinazoline-2,4(1H,3H)-dione-based emissive uracil analogue was identified to be an ideal donor for 7-diethylaminocoumarin-3-carboxylic acid. This donor/acceptor pair displays a critical Forster radius (R(0)) of 27 A, a value suitable for an A-site-aminoglycoside assembly. Titrating the coumarin labeled aminoglycosides into the emissive A-site construct, labeled at position U1406, shows a decrease in donor emission (at 395 nm) and concurrent increase of the acceptor emission (at 473 nm). Titration curves, obtained by fitting the donor's emission quenching or the augmentation of the acceptor's sensitized emission, faithfully generate EC(50) values. Titration of unlabeled ligands into the preformed FRET complex showed a continuous increase of the donor emission, with a concurrent decrease of the acceptor emission, yielding valuable data regarding competitive displacement of aminoglycosides by A-site binders. Detection of antibiotic binding is therefore not dependent on changes in the environment of a single fluorophore, but rather on the responsive interaction between two chromophores acting as a FRET pair, facilitating the determination of direct binding and competitive displacement events with FRET accuracy.
Figures

Similar articles
-
FRET Assay for Ligands Targeting the Bacterial A-Site RNA.Methods Mol Biol. 2019;1973:251-260. doi: 10.1007/978-1-4939-9216-4_16. Methods Mol Biol. 2019. PMID: 31016707
-
Confocal microscopic dual-laser dual-polarization FRET (2polFRET) at the acceptor side for correlating rotations at different distances on the cell surface.Biochim Biophys Acta Gen Subj. 2018 Apr;1862(4):1050-1068. doi: 10.1016/j.bbagen.2017.12.013. Epub 2017 Dec 30. Biochim Biophys Acta Gen Subj. 2018. PMID: 29292190
-
Modification of förster resonance energy transfer efficiencyat interfaces.Int J Mol Sci. 2012 Nov 19;13(11):15227-40. doi: 10.3390/ijms131115227. Int J Mol Sci. 2012. PMID: 23203121 Free PMC article.
-
Detecting RNA/DNA hybridization using double-labeled donor probes with enhanced fluorescence resonance energy transfer signals.Methods Mol Biol. 2006;335:43-56. doi: 10.1385/1-59745-069-3:43. Methods Mol Biol. 2006. PMID: 16785619 Review.
-
Fluorescence resonance energy transfer (FRET) and competing processes in donor-acceptor substituted DNA strands: a comparative study of ensemble and single-molecule data.J Biotechnol. 2002 Jan;82(3):211-31. doi: 10.1016/s1389-0352(01)00039-3. J Biotechnol. 2002. PMID: 11999691 Review.
Cited by
-
Interbase FRET in RNA: from A to Z.Nucleic Acids Res. 2019 Nov 4;47(19):9990-9997. doi: 10.1093/nar/gkz812. Nucleic Acids Res. 2019. PMID: 31544922 Free PMC article.
-
Interbase-FRET binding assay for pre-microRNAs.Sci Rep. 2021 Apr 30;11(1):9396. doi: 10.1038/s41598-021-88922-0. Sci Rep. 2021. PMID: 33931703 Free PMC article.
-
Site-specific fluorescent probing of RNA molecules by unnatural base-pair transcription for local structural conformation analysis.Nat Protoc. 2010 Jul;5(7):1312-23. doi: 10.1038/nprot.2010.77. Epub 2010 Jun 24. Nat Protoc. 2010. PMID: 20595959
-
Selectively guanidinylated aminoglycosides as antibiotics.ChemMedChem. 2012 Jul;7(7):1237-44. doi: 10.1002/cmdc.201200150. Epub 2012 May 25. ChemMedChem. 2012. PMID: 22639134 Free PMC article.
-
A fluorescence-based screen for ribosome binding antibiotics.Anal Biochem. 2013 Mar 15;434(2):300-7. doi: 10.1016/j.ab.2012.12.003. Epub 2012 Dec 19. Anal Biochem. 2013. PMID: 23262284 Free PMC article.
References
-
- Green R, Noller HF. Annu Rev Biochem. 1997;66:679–716. - PubMed
-
- Puglisi JD, Blanchard SC, Dahlquist KD, Eason RG, Fourmy D, Lynch SR, Recht MI, Yoshizawa S. Aminoglycosides Antibiotics and Decoding. In: Garrett RA, Douthwaite SR, Liljas A, Matheson AT, Moore PB, Noller HF, editors. The Ribosome: Structure, Function, Antibiotics, and Cellular Interactions. ASM Press; Washington, DC: 2000. pp. 419–429.
-
- Rodnina MV, Wintermeyer W. Trends Biochem Sci. 2001;26:124–130. - PubMed
-
- Gale EF, Cundliffe E, Renolds PE, Richmond MH, Waring MJ. The Molecular Basis of Antibiotic Action. John Wiley & Sons; London: 1981.
-
- Moazed D, Noller HF. Nature. 1987;327:389–394. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

