Fluorescent marker for direct detection of specific dsDNA sequences
- PMID: 19908852
- PMCID: PMC2811260
- DOI: 10.1021/ac9019895
Fluorescent marker for direct detection of specific dsDNA sequences
Abstract
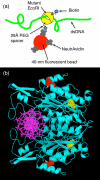
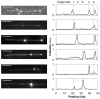
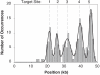
We have created a fluorescent marker using a mutant EcoRI restriction endonuclease (K249C) that enables prolonged, direct visualization of specific sequences on genomic lengths of double-stranded (ds) DNA. The marker consists of a biotinylated enzyme, attached through the biotin-avidin interaction to a fluorescent nanosphere. Control over biotin position with respect to the enzyme's binding pocket is achieved by biotinylating the mutant EcoRI at the mutation site. Biotinylated enzyme is incubated with dsDNA and NeutrAvidin-coated, fluorescent nanospheres under conditions that allow enzyme binding but prevent cleavage. Marker-laden DNA is then fluorescently stained and stretched on polylysine-coated glass slides so that the positions of the bound markers along individual DNA molecules can be measured. We demonstrate the marker's ability to bind specifically to its target sequence using both bulk gel-shift assays and single-molecule methods.
Figures


References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

