Comparison of Affymetrix Gene Array with the Exon Array shows potential application for detection of transcript isoform variation
- PMID: 19909511
- PMCID: PMC2780461
- DOI: 10.1186/1471-2164-10-519
Comparison of Affymetrix Gene Array with the Exon Array shows potential application for detection of transcript isoform variation
Abstract
Background: The emergence of isoform-sensitive microarrays has helped fuel in-depth studies of the human transcriptome. The Affymetrix GeneChip Human Exon 1.0 ST Array (Exon Array) has been previously shown to be effective in profiling gene expression at the isoform level. More recently, the Affymetrix GeneChip Human Gene 1.0 ST Array (Gene Array) has been released for measuring gene expression and interestingly contains a large subset of probes from the Exon Array. Here, we explore the potential of using Gene Array probes to assess expression variation at the sub-transcript level. Utilizing datasets of the high quality Microarray Quality Control (MAQC) RNA samples previously assayed on the Exon Array and Gene Array, we compare the expression measurements of the two platforms to determine the performance of the Gene Array in detecting isoform variations.
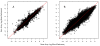
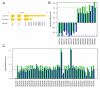
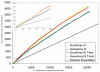
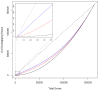
Results: Overall, we show that the Gene Array is comparable to the Exon Array in making gene expression calls. Moreover, to examine expression of different isoforms, we modify the Gene Array probe set definition file to enable summarization of probe intensity values at the exon level and show that the expression profiles between the two platforms are also highly correlated. Next, expression calls of previously known differentially spliced genes were compared and also show concordant results. Splicing index analysis, representing estimates of exon inclusion levels, shows a lower but good correlation between platforms. As the Gene Array contains a significant subset of probes from the Exon Array, we note that, in comparison, the Gene Array overlaps with fewer but still a high proportion of splicing events annotated in the Known Alt Events UCSC track, with abundant coverage of cassette exons. We discuss the ability of the Gene Array to detect alternative splicing and isoform variation and address its limitations.
Conclusion: The Gene Array is an effective expression profiling tool at gene and exon expression level, the latter made possible by probe set annotation modifications. We demonstrate that the Gene Array is capable of detecting alternative splicing and isoform variation. As expected, in comparison to the Exon Array, it is limited by reduced gene content coverage and is not able to detect as wide a range of alternative splicing events. However, for the events that can be monitored by both platforms, we estimate that the selectivity and sensitivity levels are comparable. We hope our findings will shed light on the potential extension of the Gene Array to detect alternative splicing. It should be particularly suitable for researchers primarily interested in gene expression analysis, but who may be willing to look for splicing and isoform differences within their dataset. However, we do not suggest it to be an equivalent substitute to the more comprehensive Exon Array.
Figures

References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

