Uracil-DNA glycosylase: Structural, thermodynamic and kinetic aspects of lesion search and recognition
- PMID: 19909758
- PMCID: PMC3000906
- DOI: 10.1016/j.mrfmmm.2009.10.017
Uracil-DNA glycosylase: Structural, thermodynamic and kinetic aspects of lesion search and recognition
Abstract
Uracil appears in DNA as a result of cytosine deamination and by incorporation from the dUTP pool. As potentially mutagenic and deleterious for cell regulation, uracil must be removed from DNA. The major pathway of its repair is initiated by uracil-DNA glycosylases (UNG), ubiquitously found enzymes that hydrolyze the N-glycosidic bond of deoxyuridine in DNA. This review describes the current understanding of the mechanism of uracil search and recognition by UNG. The structure of UNG proteins from several species has been solved, revealing a specific uracil-binding pocket located in a DNA-binding groove. DNA in the complex with UNG is highly distorted to allow the extrahelical recognition of uracil. Thermodynamic studies suggest that UNG binds with appreciable affinity to any DNA, mainly due to the interactions with the charged backbone. The increase in the affinity for damaged DNA is insufficient to account for the exquisite specificity of UNG for uracil. This specificity is likely to result from multistep lesion recognition process, in which normal bases are rejected at one or several pre-excision stages of enzyme-substrate complex isomerization, and only uracil can proceed to enter the active site in a catalytically competent conformation. Search for the lesion by UNG involves random sliding along DNA alternating with dissociation-association events and partial eversion of undamaged bases for initial sampling.
Copyright (c) 2009 Elsevier B.V. All rights reserved.
Conflict of interest statement
This research was supported by the Presidium of the Russian Academy of Sciences and by Russian Foundation for Basic Research. The study sponsors were not involved in the study design, collection, analysis and interpretation of data, the writing of the manuscript, or the decision to submit the manuscript for publication.
Figures

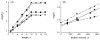

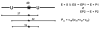
References
-
- Friedberg EC, Walker GC, Siede W, Wood RD, Schultz RA, Ellenberger T. DNA Repair and Mutagenesis. 3. ASM Press; Washington, DC: 2006.
-
- Lindahl T, Nyberg B. Heat-induced deamination of cytosine residues in deoxyribonucleic acid. Biochemistry. 1974;13:3405–3410. - PubMed
-
- Frederico LA, Kunkel TA, Shaw BR. A sensitive genetic assay for the detection of cytosine deamination: determination of rate constants and the activation energy. Biochemistry. 1990;29:2532–2537. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

