Target prediction and a statistical sampling algorithm for RNA-RNA interaction
- PMID: 19910305
- PMCID: PMC2804298
- DOI: 10.1093/bioinformatics/btp635
Target prediction and a statistical sampling algorithm for RNA-RNA interaction
Abstract
Motivation: It has been proven that the accessibility of the target sites has a critical influence on RNA-RNA binding, in general and the specificity and efficiency of miRNAs and siRNAs, in particular. Recently, O(N(6)) time and O(N(4)) space dynamic programming (DP) algorithms have become available that compute the partition function of RNA-RNA interaction complexes, thereby providing detailed insights into their thermodynamic properties.
Results: Modifications to the grammars underlying earlier approaches enables the calculation of interaction probabilities for any given interval on the target RNA. The computation of the 'hybrid probabilities' is complemented by a stochastic sampling algorithm that produces a Boltzmann weighted ensemble of RNA-RNA interaction structures. The sampling of k structures requires only negligible additional memory resources and runs in O(k.N(3)).
Availability: The algorithms described here are implemented in C as part of the rip package. The source code of rip2 can be downloaded from http://www.combinatorics.cn/cbpc/rip.html and http://www.bioinf.uni-leipzig.de/Software/rip.html.
Supplementary information: Supplementary data are available at Bioinformatics online.
Figures
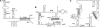




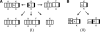



Similar articles
-
Partition function and base pairing probabilities for RNA-RNA interaction prediction.Bioinformatics. 2009 Oct 15;25(20):2646-54. doi: 10.1093/bioinformatics/btp481. Epub 2009 Aug 11. Bioinformatics. 2009. PMID: 19671692
-
Topology and prediction of RNA pseudoknots.Bioinformatics. 2011 Apr 15;27(8):1076-85. doi: 10.1093/bioinformatics/btr090. Epub 2011 Feb 17. Bioinformatics. 2011. PMID: 21335320
-
A partition function algorithm for interacting nucleic acid strands.Bioinformatics. 2009 Jun 15;25(12):i365-73. doi: 10.1093/bioinformatics/btp212. Bioinformatics. 2009. PMID: 19478011 Free PMC article.
-
Statistical and Bayesian approaches to RNA secondary structure prediction.RNA. 2006 Mar;12(3):323-31. doi: 10.1261/rna.2274106. RNA. 2006. PMID: 16495231 Free PMC article. Review.
-
How to do RNA-RNA Interaction Prediction? A Use-Case Driven Handbook Using IntaRNA.Methods Mol Biol. 2024;2726:209-234. doi: 10.1007/978-1-0716-3519-3_9. Methods Mol Biol. 2024. PMID: 38780733 Review.
Cited by
-
Analysis of CDS-located miRNA target sites suggests that they can effectively inhibit translation.Genome Res. 2013 Apr;23(4):604-15. doi: 10.1101/gr.139758.112. Epub 2013 Jan 18. Genome Res. 2013. PMID: 23335364 Free PMC article.
-
Bioinformatics of prokaryotic RNAs.RNA Biol. 2014;11(5):470-83. doi: 10.4161/rna.28647. Epub 2014 Apr 2. RNA Biol. 2014. PMID: 24755880 Free PMC article. Review.
-
An apta-aggregation based machine learning assay for rapid quantification of lysozyme through texture parameters.PLoS One. 2021 Mar 8;16(3):e0248159. doi: 10.1371/journal.pone.0248159. eCollection 2021. PLoS One. 2021. PMID: 33684138 Free PMC article.
-
A Hitchhiker's guide to RNA-RNA structure and interaction prediction tools.Brief Bioinform. 2023 Nov 22;25(1):bbad421. doi: 10.1093/bib/bbad421. Brief Bioinform. 2023. PMID: 38040490 Free PMC article. Review.
-
VfoldCPX Server: Predicting RNA-RNA Complex Structure and Stability.PLoS One. 2016 Sep 22;11(9):e0163454. doi: 10.1371/journal.pone.0163454. eCollection 2016. PLoS One. 2016. PMID: 27657918 Free PMC article.
References
-
- Akutsu T. Dynamic programming algorithms for RNA secondary structure prediction with pseudoknots. Disc. Appl. Math. 2000;104:45–62.
-
- Alkan C, et al. RNA-RNA interaction prediction and antisense RNA target search. J. Comput. Biol. 2006;13:267–282. - PubMed
-
- Andronescu M, et al. Secondary structure prediction of interacting RNA molecules. J. Mol. Biol. 2005;345:1101–1112. - PubMed
-
- Argaman L, Altuvia S. fhlA repression by OxyS RNA: kissing complex formation at two sites results in a stable antisense-target RNA complex. J. Mol. Biol. 2000;300:1101–1112. - PubMed
-
- Bachellerie J, et al. The expanding snoRNA world. Biochimie. 2002;84:775–790. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

