Histone variant H2A.Z regulates centromere silencing and chromosome segregation in fission yeast
- PMID: 19910462
- PMCID: PMC2804349
- DOI: 10.1074/jbc.M109.058487
Histone variant H2A.Z regulates centromere silencing and chromosome segregation in fission yeast
Abstract
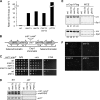
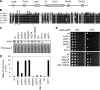
The incorporation of histone variant H2A.Z into nucleosomes plays essential roles in regulating chromatin structure and gene expression. A multisubunit complex containing chromatin remodeling protein Swr1 is responsible for the deposition of H2A.Z in budding yeast and mammals. Here, we show that the JmjC domain protein Msc1 is a novel component of the fission yeast Swr1 complex and is required for Swr1-mediated incorporation of H2A.Z into nucleosomes at gene promoters. Loss of Msc1, Swr1, or H2A.Z results in loss of silencing at centromeres and defective chromosome segregation, although centromeric levels of CENP-A, a centromere-specific histone H3 variant that is required for setting up the chromatin structure at centromeres, remain unchanged. Intriguingly, H2A.Z is required for the expression of another centromere protein, CENP-C, and overexpression of CENP-C rescues centromere silencing defects associated with H2A.Z loss. These results demonstrate the importance of H2A.Z and CENP-C in maintaining a silenced chromatin state at centromeres.
Figures




Similar articles
-
The Schizosaccharomyces pombe JmjC-protein, Msc1, prevents H2A.Z localization in centromeric and subtelomeric chromatin domains.PLoS Genet. 2009 Nov;5(11):e1000726. doi: 10.1371/journal.pgen.1000726. Epub 2009 Nov 13. PLoS Genet. 2009. PMID: 19911051 Free PMC article.
-
Heterochromatin and RNAi regulate centromeres by protecting CENP-A from ubiquitin-mediated degradation.PLoS Genet. 2018 Aug 8;14(8):e1007572. doi: 10.1371/journal.pgen.1007572. eCollection 2018 Aug. PLoS Genet. 2018. PMID: 30089114 Free PMC article.
-
The Ino80 complex mediates epigenetic centromere propagation via active removal of histone H3.Nat Commun. 2017 Sep 13;8(1):529. doi: 10.1038/s41467-017-00704-3. Nat Commun. 2017. PMID: 28904333 Free PMC article.
-
Does a GATA factor make the bed for centromeric nucleosomes?Cell Cycle. 2003 Jul-Aug;2(4):277-8. Cell Cycle. 2003. PMID: 12851470 Review.
-
Frodos found: Behold the CENP-a "Ring" bearers.Cell. 2009 May 1;137(3):409-12. doi: 10.1016/j.cell.2009.04.035. Cell. 2009. PMID: 19410539 Free PMC article. Review.
Cited by
-
Histone variants and epigenetics.Cold Spring Harb Perspect Biol. 2015 Jan 5;7(1):a019364. doi: 10.1101/cshperspect.a019364. Cold Spring Harb Perspect Biol. 2015. PMID: 25561719 Free PMC article. Review.
-
Point centromere activity requires an optimal level of centromeric noncoding RNA.Proc Natl Acad Sci U S A. 2019 Mar 26;116(13):6270-6279. doi: 10.1073/pnas.1821384116. Epub 2019 Mar 8. Proc Natl Acad Sci U S A. 2019. PMID: 30850541 Free PMC article.
-
The links between chromatin spatial organization and biological function.Biochem Soc Trans. 2013 Dec;41(6):1634-9. doi: 10.1042/BST20130213. Biochem Soc Trans. 2013. PMID: 24256267 Free PMC article. Review.
-
Histone Variants: Guardians of Genome Integrity.Cells. 2020 Nov 5;9(11):2424. doi: 10.3390/cells9112424. Cells. 2020. PMID: 33167489 Free PMC article. Review.
-
Shaping the landscape: mechanistic consequences of ubiquitin modification of chromatin.EMBO Rep. 2012 Jun 29;13(7):619-30. doi: 10.1038/embor.2012.78. EMBO Rep. 2012. PMID: 22688965 Free PMC article. Review.
References
-
- Cleveland D. W., Mao Y., Sullivan K. F. (2003) Cell 112, 407–421 - PubMed
-
- Malik H. S., Henikoff S. (2009) Cell 138, 1067–1082 - PubMed
-
- Allis C., Jenuwein T., Reinberg D. (2006) Epigenetics, Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY
-
- Black B. E., Foltz D. R., Chakravarthy S., Luger K., Woods V. L., Jr., Cleveland D. W. (2004) Nature 430, 578–582 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

