Human ribosomal protein S13 promotes gastric cancer growth through down-regulating p27(Kip1)
- PMID: 19912438
- PMCID: PMC3822796
- DOI: 10.1111/j.1582-4934.2009.00969.x
Human ribosomal protein S13 promotes gastric cancer growth through down-regulating p27(Kip1)
Abstract
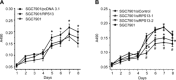
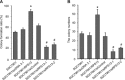
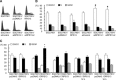
Our previous works revealed that human ribosomal protein S13 (RPS13) was up-regulated in multidrug-resistant gastric cancer cells and overexpression of RPS13 could protect gastric cancer cells from drug-induced apoptosis. The present study was designed to explore the role of RPS13 in tumorigenesis and development of gastric cancer. The expression of RPS13 in gastric cancer tissues and normal gastric mucosa was evaluated by immunohistochemical staining and Western blot analysis. It was found RPS13 was expressed at a higher level in gastric cancer tissues than that in normal gastric mucosa. RPS13 was then genetically overexpressed in gastric cancer cells or knocked down by RNA interference. It was demonstrated that up-regulation of RPS13 accelerated the growth, enhanced in vitro colony forming and soft agar cologenic ability and promoted in vivo tumour formation potential of gastric cancer cells. Meanwhile, down-regulation of RPS13 in gastric cancer cells resulted in complete opposite effects. Moreover, overexpression of RPS13 could promote G1 to S phase transition whereas knocking down of RPS13 led to G1 arrest of gastric cancer cells. It was further demonstrated that RPS13 down-regulated p27(kip1) expression and CDK2 kinase activity but did not change the expression of cyclin D, cyclin E, CDK2, CDK4 and p16(INK4A). Taken together, these data indicate that RPS13 could promote the growth and cell cycle progression of gastric cancer cells at least through inhibiting p27(kip1) expression.
© 2011 The Authors Journal of Cellular and Molecular Medicine © 2011 Foundation for Cellular and Molecular Medicine/Blackwell Publishing Ltd.
Figures




References
-
- Denis MG, Chadeneau C, Lecabellec MT, et al. Over-expression of the S13 riboso-mal protein in actively growing cells. Int J Cancer. 1993;55:275–80. - PubMed
-
- Zhang L, Cilley RE, Chinoy MR. Suppression subtractive hybridization to identify gene expressions in variant and classic small cell lung cancer cell lines. J Surg Res. 2000;93:108–19. - PubMed
-
- Kondoh N, Wakatsuki T, Ryo A, et al. Identification and characterization of genes associated with human hepatocellular car-cinogenesis. Cancer Res. 1999;59:4990–6. - PubMed
-
- Ganger DR, Hamilton PD, Klos DJ, et al. Differential expression of metallopanstim-ulin/S27 ribosomal protein in hepatic regeneration and neoplasia. Cancer Detect Prev. 2001;25:231–6. - PubMed
-
- Vaarala MH, Porvari KS, Kyllönen AP, et al. Several genes encoding ribosomal proteins are over-expressed in prostate-cancer cell lines: confirmation of L7a and L37 over-expression in prostate-cancer tissue samples. Int J Cancer. 1998;78:27–32. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

