Genome wide evolutionary analyses reveal serotype specific patterns of positive selection in selected Salmonella serotypes
- PMID: 19912661
- PMCID: PMC2784778
- DOI: 10.1186/1471-2148-9-264
Genome wide evolutionary analyses reveal serotype specific patterns of positive selection in selected Salmonella serotypes
Abstract
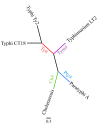
Background: The bacterium Salmonella enterica includes a diversity of serotypes that cause disease in humans and different animal species. Some Salmonella serotypes show a broad host range, some are host restricted and exclusively associated with one particular host, and some are associated with one particular host species, but able to cause disease in other host species and are thus considered "host adapted". Five Salmonella genome sequences, representing a broad host range serotype (Typhimurium), two host restricted serotypes (Typhi [two genomes] and Paratyphi) and one host adapted serotype (Choleraesuis) were used to identify core genome genes that show evidence for recombination and positive selection.
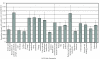
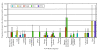
Results: Overall, 3323 orthologous genes were identified in all 5 Salmonella genomes analyzed. Use of four different methods to assess homologous recombination identified 270 genes that showed evidence for recombination with at least one of these methods (false discovery rate [FDR] <10%). After exclusion of genes with evidence for recombination, site and branch specific models identified 41 genes as showing evidence for positive selection (FDR <20%), including a number of genes with confirmed or likely roles in virulence and ompC, a gene encoding an outer membrane protein, which has also been found to be under positive selection in other bacteria. A total of 8, 16, 7, and 5 genes showed evidence for positive selection in Choleraesuis, Typhi, Typhimurium, and Paratyphi branch analyses, respectively. Sequencing and evolutionary analyses of four genes in an additional 42 isolates representing 23 serotypes confirmed branch specific positive selection and recombination patterns.
Conclusion: Our data show that, among the four serotypes analyzed, (i) less than 10% of Salmonella genes in the core genome show evidence for homologous recombination, (ii) a number of Salmonella genes are under positive selection, including genes that appear to contribute to virulence, and (iii) branch specific positive selection contributes to the evolution of host restricted Salmonella serotypes.
Figures
Similar articles
-
Serotype-specific evolutionary patterns of antimicrobial-resistant Salmonella enterica.BMC Evol Biol. 2019 Jun 21;19(1):132. doi: 10.1186/s12862-019-1457-5. BMC Evol Biol. 2019. PMID: 31226931 Free PMC article.
-
Comparative genomic analysis uncovers 3 novel loci encoding type six secretion systems differentially distributed in Salmonella serotypes.BMC Genomics. 2009 Aug 4;10:354. doi: 10.1186/1471-2164-10-354. BMC Genomics. 2009. PMID: 19653904 Free PMC article.
-
Genome-wide analyses reveal lineage specific contributions of positive selection and recombination to the evolution of Listeria monocytogenes.BMC Evol Biol. 2008 Aug 12;8:233. doi: 10.1186/1471-2148-8-233. BMC Evol Biol. 2008. PMID: 18700032 Free PMC article.
-
[Evolutionary engineering in Salmonella: emergence of hybrid virulence-resistance plasmids in non-typhoid serotypes].Enferm Infecc Microbiol Clin. 2009 Jan;27(1):37-43. doi: 10.1016/j.eimc.2008.09.001. Epub 2009 Jan 9. Enferm Infecc Microbiol Clin. 2009. PMID: 19218002 Review. Spanish.
-
Salmonella: a model for bacterial pathogenesis.Annu Rev Med. 2001;52:259-74. doi: 10.1146/annurev.med.52.1.259. Annu Rev Med. 2001. PMID: 11160778 Review.
Cited by
-
Microfluidic PCR combined with pyrosequencing for identification of allelic variants with phenotypic associations among targeted Salmonella genes.Appl Environ Microbiol. 2012 Oct;78(20):7480-2. doi: 10.1128/AEM.01703-12. Epub 2012 Aug 10. Appl Environ Microbiol. 2012. PMID: 22885744 Free PMC article.
-
Identification of positively selected genes in human pathogenic treponemes: Syphilis-, yaws-, and bejel-causing strains differ in sets of genes showing adaptive evolution.PLoS Negl Trop Dis. 2019 Jun 19;13(6):e0007463. doi: 10.1371/journal.pntd.0007463. eCollection 2019 Jun. PLoS Negl Trop Dis. 2019. PMID: 31216284 Free PMC article.
-
Phenotypic and Genotypic Eligible Methods for Salmonella Typhimurium Source Tracking.Front Microbiol. 2017 Dec 22;8:2587. doi: 10.3389/fmicb.2017.02587. eCollection 2017. Front Microbiol. 2017. PMID: 29312260 Free PMC article. Review.
-
A genome-wide identification of genes undergoing recombination and positive selection in Neisseria.Biomed Res Int. 2014;2014:815672. doi: 10.1155/2014/815672. Epub 2014 Aug 10. Biomed Res Int. 2014. PMID: 25180194 Free PMC article.
-
PosiGene: automated and easy-to-use pipeline for genome-wide detection of positively selected genes.Nucleic Acids Res. 2017 Jun 20;45(11):e100. doi: 10.1093/nar/gkx179. Nucleic Acids Res. 2017. PMID: 28334822 Free PMC article.
References
-
- McClelland M, Sanderson KE, Clifton SW, Latreille P, Porwollik S, Sabo A, Meyer R, Bieri T, Ozersky P, McLellan M, Harkins CR, Wang CY, Nguyen C, Berghoff A, Elliott G, Kohlberg S, Strong C, Du FY, Carter J, Kremizki C, Layman D, Leonard S, Sun H, Fulton L, Nash W, Miner T, Minx P, Delehaunty K, Fronick C, Magrini V, Nhan M, Warren W, Florea L, Spieth J, Wilson RK. Comparison of genome degradation in Paratyphi A and Typhi, human-restricted serovars of Salmonella enterica that cause typhoid. Nat Gen. 2004;36(12):1268–1274. doi: 10.1038/ng1470. - DOI - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

