The structural biochemistry of the superoxide dismutases
- PMID: 19914407
- PMCID: PMC3098211
- DOI: 10.1016/j.bbapap.2009.11.004
The structural biochemistry of the superoxide dismutases
Abstract
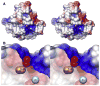
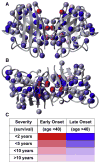
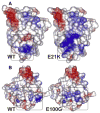
The discovery of superoxide dismutases (SODs), which convert superoxide radicals to molecular oxygen and hydrogen peroxide, has been termed the most important discovery of modern biology never to win a Nobel Prize. Here, we review the reasons this discovery has been underappreciated, as well as discuss the robust results supporting its premier biological importance and utility for current research. We highlight our understanding of SOD function gained through structural biology analyses, which reveal important hydrogen-bonding schemes and metal-binding motifs. These structural features create remarkable enzymes that promote catalysis at faster than diffusion-limited rates by using electrostatic guidance. These architectures additionally alter the redox potential of the active site metal center to a range suitable for the superoxide disproportionation reaction and protect against inhibition of catalysis by molecules such as phosphate. SOD structures may also control their enzymatic activity through product inhibition; manipulation of these product inhibition levels has the potential to generate therapeutic forms of SOD. Markedly, structural destabilization of the SOD architecture can lead to disease, as mutations in Cu,ZnSOD may result in familial amyotrophic lateral sclerosis, a relatively common, rapidly progressing and fatal neurodegenerative disorder. We describe our current understanding of how these Cu,ZnSOD mutations may lead to aggregation/fibril formation, as a detailed understanding of these mechanisms provides new avenues for the development of therapeutics against this so far untreatable neurodegenerative pathology.
Copyright 2009 Elsevier B.V. All rights reserved.
Figures

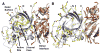
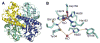
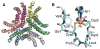
References
-
- McCord JM, Fridovich I. Superoxide dismutase. An enzymic function for erythrocuprein (hemocuprein) J Biol Chem. 1969;244:6049–6055. - PubMed
-
- Fridovich I. Superoxide anion radical (O2-.), superoxide dismutases, and related matters. J Biol Chem. 1997;272:18515–18517. - PubMed
-
- Lane N. Oxygen: The Molecule that Made the World. Oxford University Press; USA: 2003.
-
- Fridovich I. Superoxide dismutases. Adv Enzymol Relat Areas Mol Biol. 1986;58:61–97. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

