Recurrent reciprocal 16p11.2 rearrangements associated with global developmental delay, behavioural problems, dysmorphism, epilepsy, and abnormal head size
- PMID: 19914906
- PMCID: PMC3158566
- DOI: 10.1136/jmg.2009.073015
Recurrent reciprocal 16p11.2 rearrangements associated with global developmental delay, behavioural problems, dysmorphism, epilepsy, and abnormal head size
Abstract
Background: Deletion and the reciprocal duplication in 16p11.2 were recently associated with autism and developmental delay.
Method: We indentified 27 deletions and 18 duplications of 16p11.2 were identified in 0.6% of all samples submitted for clinical array-CGH (comparative genomic hybridisation) analysis. Detailed molecular and phenotypic characterisations were performed on 17 deletion subjects and ten subjects with the duplication.
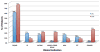
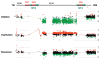
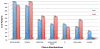
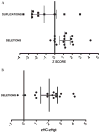
Results: The most common clinical manifestations in 17 deletion and 10 duplication subjects were speech/language delay and cognitive impairment. Other phenotypes in the deletion patients included motor delay (50%), seizures ( approximately 40%), behavioural problems ( approximately 40%), congenital anomalies ( approximately 30%), and autism ( approximately 20%). The phenotypes among duplication patients included motor delay (6/10), behavioural problems (especially attention deficit hyperactivity disorder (ADHD)) (6/10), congenital anomalies (5/10), and seizures (3/10). Patients with the 16p11.2 deletion had statistically significant macrocephaly (p<0.0017) and 6 of the 10 patients with the duplication had microcephaly. One subject with the deletion was asymptomatic and another with the duplication had a normal cognitive and behavioural phenotype. Genomic analyses revealed additional complexity to the 16p11.2 region with mechanistic implications. The chromosomal rearrangement was de novo in all but 2 of the 10 deletion cases in which parental studies were available. Additionally, 2 de novo cases were apparently mosaic for the deletion in the analysed blood sample. Three de novo and 2 inherited cases were observed in the 5 of 10 duplication patients where data were available.
Conclusions: Recurrent reciprocal 16p11.2 deletion and duplication are characterised by a spectrum of primarily neurocognitive phenotypes that are subject to incomplete penetrance and variable expressivity. The autism and macrocephaly observed with deletion and ADHD and microcephaly seen in duplication patients support a diametric model of autism spectrum and psychotic spectrum behavioural phenotypes in genomic sister disorders.
Conflict of interest statement
Figures



References
-
- Shinawi M, Cheung SW. The array CGH and its clinical applications. Drug Discov Today. 2008;13:760–70. - PubMed
-
- Menten B, Maas N, Thienpont B, Buysse K, Vandesompele J, Melotte C, de Ravel T, Van Vooren S, Balikova I, Backx L, Janssens S, De Paepe A, De Moor B, Moreau Y, Marynen P, Fryns JP, Mortier G, Devriendt K, Speleman F, Vermeesch JR. Emerging patterns of cryptic chromosomal imbalances in patients with idiopathic mental retardation and multiple congenital anomalies: a new series of 140 patients and review of the literature. J Med Genet. 2006;43:625–33. - PMC - PubMed
-
- Jacquemont ML, Sanlaville D, Redon R, Raoul O, Cormier-Daire V, Lyonnet S, Amiel J, Le Merrer M, Heron D, de Blois MC, Prieur M, Vekemans M, Carter NP, Munnich A, Colleaux L, Philippe A. Abased comparative genomic hybridisation identifies high frequency of cryptic chromosomal rearrangements in patients with syndromic autism spectrum disorders. J Med Genet. 2006;43:843–9. - PMC - PubMed
-
- Sebat J, Lakshmi B, Malhotra D, Troge J, Lese-Martin C, Walsh T, Yamrom B, Yoon S, Krasnitz A, Kendall J, Leotta A, Pai D, Zhang R, Lee YH, Hicks J, Spence SJ, Lee AT, Puura K, Lehtimäki T, Ledbetter D, Gregersen PK, Bregman J, Sutcliffe JS, Jobanputra V, Chung W, Warburton D, King MC, Skuse D, Geschwind DH, Gilliam TC, Ye K, Wigler M. Strong association of de novo copy number mutations with autism. Science. 2007;316:445–9. - PMC - PubMed
-
- Marshall CR, Noor A, Vincent JB, Lionel AC, Feuk L, Skaug J, Shago M, Moessner R, Pinto D, Ren Y, Thiruvahindrapduram B, Fiebig A, Schreiber S, Friedman J, Ketelaars CE, Vos YJ, Ficicioglu C, Kirkpatrick S, Nicolson R, Sloman L, Summers A, Gibbons CA, Teebi A, Chitayat D, Weksberg R, Thompson A, Vardy C, Crosbie V, Luscombe S, Baatjes R, Zwaigenbaum L, Roberts W, Fernandez B, Szatmari P, Scherer SW. Structural variation of chromosomes in autism spectrum disorder. Am J Hum Genet. 2008;82:477–88. - PMC - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
