Probing the (H3-H4)2 histone tetramer structure using pulsed EPR spectroscopy combined with site-directed spin labelling
- PMID: 19914933
- PMCID: PMC2810997
- DOI: 10.1093/nar/gkp1003
Probing the (H3-H4)2 histone tetramer structure using pulsed EPR spectroscopy combined with site-directed spin labelling
Abstract
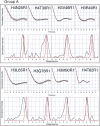
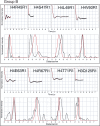
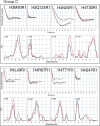
The (H3-H4)(2) histone tetramer forms the central core of nucleosomes and, as such, plays a prominent role in assembly, disassembly and positioning of nucleosomes. Despite its fundamental role in chromatin, the tetramer has received little structural investigation. Here, through the use of pulsed electron-electron double resonance spectroscopy coupled with site-directed spin labelling, we survey the structure of the tetramer in solution. We find that tetramer is structurally more heterogeneous on its own than when sequestered in the octamer or nucleosome. In particular, while the central region including the H3-H3' interface retains a structure similar to that observed in nucleosomes, other regions such as the H3 alphaN helix display increased structural heterogeneity. Flexibility of the H3 alphaN helix in the free tetramer also illustrates the potential for post-translational modifications to alter the structure of this region and mediate interactions with histone chaperones. The approach described here promises to prove a powerful system for investigating the structure of additional assemblies of histones with other important factors in chromatin assembly/fluidity.
Figures


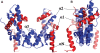
References
-
- Luger K, Rechsteiner TJ, Flaus AJ, Waye MM, Richmond TJ. Characterization of nucleosome core particles containing histone proteins made in bacteria. J. Mol. Biol. 1997;272:301–311. - PubMed
-
- Jorcano JL, Ruiz-Carrillo A. H3.H4 tetramer directs DNA and core histone octamer assembly in the nucleosome core particle. Biochemistry. 1979;18:768–774. - PubMed
-
- Polach KJ, Widom J. Mechanism of protein access to specific DNA-sequences in chromatin – a dynamic equilibrium-model for gene-regulation. J. Mol. Biol. 1995;254:130–149. - PubMed

