The missing link in linear alkylbenzenesulfonate surfactant degradation: 4-sulfoacetophenone as a transient intermediate in the degradation of 3-(4-sulfophenyl)butyrate by Comamonas testosteroni KF-1
- PMID: 19915037
- PMCID: PMC2798632
- DOI: 10.1128/AEM.02181-09
The missing link in linear alkylbenzenesulfonate surfactant degradation: 4-sulfoacetophenone as a transient intermediate in the degradation of 3-(4-sulfophenyl)butyrate by Comamonas testosteroni KF-1
Abstract
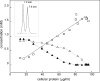
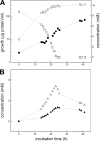
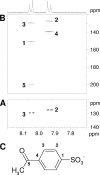
Biodegradation of the laundry surfactant linear alkylbenzenesulfonate (LAS) involves complex bacterial communities. The known heterotrophic community has two tiers. First, all LAS congeners are oxygenated and oxidized to about 50 sulfophenylcarboxylates (SPC). Second, the SPCs are mineralized. Comamonas testosteroni KF-1 mineralizes 3-(4-sulfophenyl)butyrate (3-C4-SPC). During growth of strain KF-1 with 3-C4-SPC, two transient intermediates were detected in the culture medium. One intermediate was identified as 4-sulfoacetophenone (SAP) (4-acetylbenzenesulfonate) by nuclear magnetic resonance (NMR). The other was 4-sulfophenol (SP). This information allowed us to postulate a degradation pathway that comprises the removal of an acetyl moiety from (derivatized) 3-C4-SPC, followed by a Baeyer-Villiger monooxygenation of SAP and subsequent ester cleavage to yield SP. Inducible NADPH-dependent SAP-oxygenase was detected in crude extracts of strain KF-1. The enzyme reaction involved transient formation of 4-sulfophenol acetate (SPAc), which was completely hydrolyzed to SP and acetate. SP was subject to NADH-dependent oxygenation in crude extract, and 4-sulfocatechol (SC) was subject to oxygenolytic ring cleavage. The first complete degradative pathway for an SPC can now be depicted with 3-C4-SPC: transport, ligation to a coenzyme A (CoA) ester, and manipulation to allow abstraction of acetyl-CoA to yield SAP, Baeyer-Villiger monooxygenation to SPAc, hydrolysis of the ester to acetate and SP, monooxygenation of SP to SC, the ortho ring-cleavage pathway with desulfonation, and sulfite oxidation.
Figures

Similar articles
-
Two enzymes of a complete degradation pathway for linear alkylbenzenesulfonate (LAS) surfactants: 4-sulfoacetophenone Baeyer-Villiger monooxygenase and 4-sulfophenylacetate esterase in Comamonas testosteroni KF-1.Appl Environ Microbiol. 2012 Dec;78(23):8254-63. doi: 10.1128/AEM.02412-12. Epub 2012 Sep 21. Appl Environ Microbiol. 2012. PMID: 23001656 Free PMC article.
-
Mineralization of individual congeners of linear alkylbenzenesulfonate by defined pairs of heterotrophic bacteria.Appl Environ Microbiol. 2004 Jul;70(7):4053-63. doi: 10.1128/AEM.70.7.4053-4063.2004. Appl Environ Microbiol. 2004. PMID: 15240283 Free PMC article.
-
Parvibaculum lavamentivorans DS-1T degrades centrally substituted congeners of commercial linear alkylbenzenesulfonate to sulfophenyl carboxylates and sulfophenyl dicarboxylates.Appl Environ Microbiol. 2007 Aug;73(15):4725-32. doi: 10.1128/AEM.00632-07. Epub 2007 Jun 8. Appl Environ Microbiol. 2007. PMID: 17557839 Free PMC article.
-
Omega-oxygenation of the alkyl sidechain of linear alkylbenzenesulfonate (LAS) surfactant in Parvibaculum lavamentivorans(T).Arch Microbiol. 2005 Sep;183(6):369-77. doi: 10.1007/s00203-005-0002-7. Epub 2005 Oct 13. Arch Microbiol. 2005. PMID: 16075201
-
Alpha,beta-unsaturated sulfophenylcarboxylates as degradation intermediates of linear alkylbenzenesulfonates: evidence for omega-oxygenation followed by beta-oxidations by liquid chromatography-mass spectrometry.Environ Toxicol Chem. 2002 Jan;21(1):1-8. Environ Toxicol Chem. 2002. PMID: 11804041
Cited by
-
Two enzymes of a complete degradation pathway for linear alkylbenzenesulfonate (LAS) surfactants: 4-sulfoacetophenone Baeyer-Villiger monooxygenase and 4-sulfophenylacetate esterase in Comamonas testosteroni KF-1.Appl Environ Microbiol. 2012 Dec;78(23):8254-63. doi: 10.1128/AEM.02412-12. Epub 2012 Sep 21. Appl Environ Microbiol. 2012. PMID: 23001656 Free PMC article.
-
Comamonas testosteronan synthase, a bifunctional glycosyltransferase that produces a unique heparosan polysaccharide analog.Glycobiology. 2011 Oct;21(10):1331-40. doi: 10.1093/glycob/cwr072. Epub 2011 May 24. Glycobiology. 2011. PMID: 21610195 Free PMC article.
-
Permanent draft genome sequence of Comamonas testosteroni KF-1.Stand Genomic Sci. 2013 May 30;8(2):239-54. doi: 10.4056/sigs.3847890. eCollection 2013. Stand Genomic Sci. 2013. PMID: 23991256 Free PMC article.
-
Composition of "gold juice" using an ancient method based on intestinal microecology.J Int Med Res. 2020 Sep;48(9):300060520931288. doi: 10.1177/0300060520931288. J Int Med Res. 2020. PMID: 32993381 Free PMC article.
References
-
- Cain, R. B. 1987. Biodegradation of anionic surfactants. Biochem. Soc. Trans. 15:7-22. - PubMed
-
- Contzen, M., S. Bürger, and A. Stolz. 2001. Cloning of the genes for a 4-sulphocatechol-oxidizing protocatechuate 3,4-dioxygenase from Hydrogenophaga intermedia S1 and identification of the amino acid residues responsible for the ability to convert 4-sulphocatechol. Mol. Microbiol. 41:199-205. - PubMed
-
- Darby, J. M., D. G. Taylor, and D. J. Hopper. 1987. Hydroquinone as the ring-fission substrate in the catabolism of 4-ethylphenol and 4-hydroxyacetophenone by Pseudomonas putida JD1. J. Gen. Microbiol. 133:2137-2146.
-
- Denger, K., S. Weinitschke, T. H. Smits, D. Schleheck, and A. M. Cook. 2008. Bacterial sulfite dehydrogenases in organotrophic metabolism: separation and identification in Cupriavidus necator H16 and in Delftia acidovorans SPH-1. Microbiology 154:256-263. - PubMed
-
- Dodgson, K. S., and F. F. White. 1983. Some microbial enzymes involved in the biodegradation of sulphated surfactants. Top. Enzyme Ferment. Biotechnol. 7:90-155.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

