T cell intrinsic heterodimeric complexes between HVEM and BTLA determine receptivity to the surrounding microenvironment
- PMID: 19915044
- PMCID: PMC2865256
- DOI: 10.4049/jimmunol.0902490
T cell intrinsic heterodimeric complexes between HVEM and BTLA determine receptivity to the surrounding microenvironment
Abstract
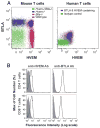
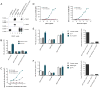
The inhibitory cosignaling pathway formed between the TNF receptor herpesvirus entry mediator (HVEM, TNFRSF14) and the Ig superfamily members, B and T lymphocyte attenuator (BTLA) and CD160, limits the activation of T cells. However, BTLA and CD160 can also serve as activating ligands for HVEM when presented in trans by adjacent cells, thus forming a bidirectional signaling pathway. BTLA and CD160 can directly activate the HVEM-dependent NF-kappaB RelA transcriptional complex raising the question of how NF-kappaB activation is repressed in naive T cells. In this study, we show BTLA interacts with HVEM in cis, forming a heterodimeric complex in naive T cells that inhibits HVEM-dependent NF-kappaB activation. The cis-interaction between HVEM and BTLA is the predominant form expressed on the surface of naive human and mouse T cells. The BTLA ectodomain acts as a competitive inhibitor blocking BTLA and CD160 from binding in trans to HVEM and initiating NF-kappaB activation. The TNF-related ligand, LIGHT (homologous to lymphotoxins, exhibits inducible expression, and competes with HSV glycoprotein D for HVEM, a receptor expressed by T lymphocytes, or TNFSF14) binds HVEM in the cis-complex, but NF-kappaB activation was attenuated, suggesting BTLA prevents oligomerization of HVEM in the cis-complex. Genetic deletion of BTLA or pharmacologic disruption of the HVEM-BTLA cis-complex in T cells promoted HVEM activation in trans. Interestingly, herpes simplex virus envelope glycoprotein D formed a cis-complex with HVEM, yet surprisingly, promoted the activation NF-kappaB RelA. We suggest that the HVEM-BTLA cis-complex competitively inhibits HVEM activation by ligands expressed in the surrounding microenvironment, thus helping maintain T cells in the naive state.
Conflict of interest statement
The authors have no financial conflict of interest.
Figures




Similar articles
-
Unconventional ligand activation of herpesvirus entry mediator signals cell survival.Proc Natl Acad Sci U S A. 2009 Apr 14;106(15):6244-9. doi: 10.1073/pnas.0902115106. Epub 2009 Mar 30. Proc Natl Acad Sci U S A. 2009. PMID: 19332782 Free PMC article.
-
Molecular basis for herpesvirus entry mediator recognition by the human immune inhibitory receptor CD160 and its relationship to the cosignaling molecules BTLA and LIGHT.J Mol Biol. 2011 Nov 4;413(4):762-72. doi: 10.1016/j.jmb.2011.09.018. Epub 2011 Sep 19. J Mol Biol. 2011. PMID: 21959263
-
The CD160, BTLA, LIGHT/HVEM pathway: a bidirectional switch regulating T-cell activation.Immunol Rev. 2009 May;229(1):244-58. doi: 10.1111/j.1600-065X.2009.00783.x. Immunol Rev. 2009. PMID: 19426226 Review.
-
TNF Superfamily Networks: bidirectional and interference pathways of the herpesvirus entry mediator (TNFSF14).Curr Opin Immunol. 2011 Oct;23(5):627-31. doi: 10.1016/j.coi.2011.08.008. Epub 2011 Sep 13. Curr Opin Immunol. 2011. PMID: 21920726 Free PMC article. Review.
-
A herpesvirus entry mediator mutein with selective agonist action for the inhibitory receptor B and T lymphocyte attenuator.J Biol Chem. 2017 Dec 22;292(51):21060-21070. doi: 10.1074/jbc.M117.813295. Epub 2017 Oct 23. J Biol Chem. 2017. PMID: 29061848 Free PMC article.
Cited by
-
Selective blockade of herpesvirus entry mediator-B and T lymphocyte attenuator pathway ameliorates acute graft-versus-host reaction.J Immunol. 2012 May 15;188(10):4885-96. doi: 10.4049/jimmunol.1103698. Epub 2012 Apr 6. J Immunol. 2012. PMID: 22490863 Free PMC article.
-
Redesigning HVEM Interface for Selective Binding to LIGHT, BTLA, and CD160.Structure. 2020 Nov 3;28(11):1197-1205.e2. doi: 10.1016/j.str.2020.07.013. Epub 2020 Aug 13. Structure. 2020. PMID: 32795404 Free PMC article.
-
Immune checkpoint signaling and cancer immunotherapy.Cell Res. 2020 Aug;30(8):660-669. doi: 10.1038/s41422-020-0343-4. Epub 2020 May 28. Cell Res. 2020. PMID: 32467592 Free PMC article. Review.
-
The HVEM-BTLA Axis Restrains T Cell Help to Germinal Center B Cells and Functions as a Cell-Extrinsic Suppressor in Lymphomagenesis.Immunity. 2019 Aug 20;51(2):310-323.e7. doi: 10.1016/j.immuni.2019.05.022. Epub 2019 Jun 13. Immunity. 2019. PMID: 31204070 Free PMC article.
-
PD-L1 Binds to B7-1 Only In Cis on the Same Cell Surface.Cancer Immunol Res. 2018 Aug;6(8):921-929. doi: 10.1158/2326-6066.CIR-17-0316. Epub 2018 Jun 5. Cancer Immunol Res. 2018. PMID: 29871885 Free PMC article.
References
-
- Montgomery RI, Warner MS, Lum B, Spear PG. Herpes simplex virus 1 entry into cells mediated by a novel member of the TNF/NGF receptor family. Cell. 1996;87:427–436. - PubMed
-
- Sedy JR, Gavrieli M, Potter KG, Hurchla MA, Lindsley RC, Hildner K, Scheu S, Pfeffer K, Ware CF, Murphy TL, Murphy KM. B and T lymphocyte attenuator regulates T cell activation through interaction with herpesvirus entry mediator. Nat Immunol. 2005;6:90–98. - PubMed
-
- Mauri DN, Ebner R, Montgomery RI, Kochel KD, Cheung TC, Yu GL, Ruben S, Murphy M, Eisenberg RJ, Cohen GH, et al. LIGHT, a new member of the TNF superfamily, and lymphotoxin α are ligands for herpesvirus entry mediator. Immunity. 1998;8:21–30. - PubMed
-
- Cai G, Anumanthan A, Brown JA, Greenfield EA, Zhu B, Freeman GJ. CD160 inhibits activation of human CD4+ T cells through interaction with herpesvirus entry mediator. Nat Immunol. 2008;9:176–185. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- AI067890/AI/NIAID NIH HHS/United States
- R24 DK080506/DK/NIDDK NIH HHS/United States
- R37AI33068/AI/NIAID NIH HHS/United States
- R37AI036293/AI/NIAID NIH HHS/United States
- R01 AI061516/AI/NIAID NIH HHS/United States
- CA069381/CA/NCI NIH HHS/United States
- R37 AI033068/AI/NIAID NIH HHS/United States
- U19 AI031494/AI/NIAID NIH HHS/United States
- AI61516/AI/NIAID NIH HHS/United States
- R01 AI067890/AI/NIAID NIH HHS/United States
- R37 AI036293/AI/NIAID NIH HHS/United States
- R01 AI048073/AI/NIAID NIH HHS/United States
- DK080506/DK/NIDDK NIH HHS/United States
- U19AI031494/AI/NIAID NIH HHS/United States
- AI048073/AI/NIAID NIH HHS/United States
- P01 CA069381/CA/NCI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

