RNA interference silencing of a major lipid droplet protein affects lipid droplet size in Chlamydomonas reinhardtii
- PMID: 19915074
- PMCID: PMC2805299
- DOI: 10.1128/EC.00203-09
RNA interference silencing of a major lipid droplet protein affects lipid droplet size in Chlamydomonas reinhardtii
Abstract
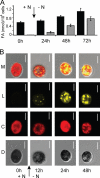
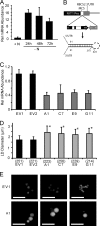
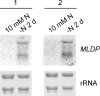
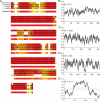
Eukaryotic cells store oils in the chemical form of triacylglycerols in distinct organelles, often called lipid droplets. These dynamic storage compartments have been intensely studied in the context of human health and also in plants as a source of vegetable oils for human consumption and for chemical or biofuel feedstocks. Many microalgae accumulate oils, particularly under conditions limiting to growth, and thus have gained renewed attention as a potentially sustainable feedstock for biofuel production. However, little is currently known at the cellular or molecular levels with regard to oil accumulation in microalgae, and the structural proteins and enzymes involved in the biogenesis, maintenance, and degradation of algal oil storage compartments are not well studied. Focusing on the model green alga Chlamydomonas reinhardtii, the accumulation of triacylglycerols and the formation of lipid droplets during nitrogen deprivation were investigated. Mass spectrometry identified 259 proteins in a lipid droplet-enriched fraction, among them a major protein, tentatively designated major lipid droplet protein (MLDP). This protein is specific to the green algal lineage of photosynthetic organisms. Repression of MLDP gene expression using an RNA interference approach led to increased lipid droplet size, but no change in triacylglycerol content or metabolism was observed.
Figures


References
-
- Abramoff, M. D., P. J. Magelhaes, and S. J. Ram. 2004. Image processing with ImageJ. Biophotonics Int. 11:36-42.
-
- Atteia, A., A. Adrait, S. Brugiere, M. Tardif, L. R. van, O. Deusch, T. Dagan, L. Kuhn, B. Gontero, W. Martin, J. Garin, J. Joyard, and N. Rolland. 2009. A proteomic survey of Chlamydomonas reinhardtii mitochondria sheds new light on the metabolic plasticity of the organelle and on the nature of the alpha-proteobacterial mitochondrial ancestor. Mol. Biol. Evol. 26:1533-1548. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

