The BTB/POZ domain of the Arabidopsis disease resistance protein NPR1 interacts with the repression domain of TGA2 to negate its function
- PMID: 19915088
- PMCID: PMC2798319
- DOI: 10.1105/tpc.109.069971
The BTB/POZ domain of the Arabidopsis disease resistance protein NPR1 interacts with the repression domain of TGA2 to negate its function
Abstract
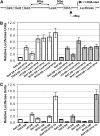
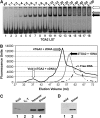
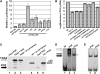
TGA2 and NONEXPRESSER OF PR GENES1 (NPR1) are activators of systemic acquired resistance (SAR) and of the SAR marker gene pathogenesis-related-1 (PR-1) in Arabidopsis thaliana. TGA2 is a transcriptional repressor required for basal repression of PR-1, but during SAR, TGA2 recruits NPR1 as part of an enhanceosome. Transactivation by the enhanceosome requires the NPR1 BTB/POZ domain. However, the NPR1 BTB/POZ domain does not contain an autonomous transactivation domain; thus, its molecular role within the enhanceosome remains elusive. We now show by gel filtration analyses that TGA2 binds DNA as a dimer, tetramer, or oligomer. Using in vivo plant transcription assays, we localize the repression domain of TGA2 to the N terminus and demonstrate that this domain is responsible for modulating the DNA binding activity of the oligomer both in vitro and in vivo. We confirm that the NPR1 BTB/POZ domain interacts with and negates the molecular function of the TGA2 repression domain by excluding TGA2 oligomers from cognate DNA. These data distinguish the NPR1 BTB/POZ domain from other known BTB/POZ domains and establish its molecular role in the context of the Arabidopsis PR-1 gene enhanceosome.
Figures




References
-
- Aoki, K., Suzuki, K., Ishida, R., and Kasai, M. (1999). The DNA-binding activity of Translin is mediated by a basic region in the ring-shaped structure conserved in evolution. FEBS Lett. 443 363–366. - PubMed
-
- Aravind, L., and Koonin, E.V. (1999). Fold prediction and evolutionary analysis of the POZ domain: Structural and evolutionary relationship with the potassium channel tetramerization domain. J. Mol. Biol. 285 1353–1361. - PubMed
-
- Bardwell, V.J., and Treisman, R. (1994). The POZ domain: A conserved protein-protein interaction motif. Genes Dev. 8 1664–1677. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

