Dof5.6/HCA2, a Dof transcription factor gene, regulates interfascicular cambium formation and vascular tissue development in Arabidopsis
- PMID: 19915089
- PMCID: PMC2798324
- DOI: 10.1105/tpc.108.064139
Dof5.6/HCA2, a Dof transcription factor gene, regulates interfascicular cambium formation and vascular tissue development in Arabidopsis
Abstract
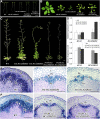
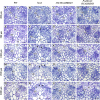
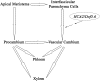
Vascular cambium, a type of lateral meristem, is the source of secondary xylem and secondary phloem, but little is known about the molecular mechanisms of its formation and development. Here, we report the characterization of an Arabidopsis thaliana gain-of-function mutant with dramatically increased cambial activity, designated high cambial activity2 (hca2). The hca2 mutant has no alternative organization of the vascular bundles/fibers in inflorescence stems, due to precocious formation of interfascicular cambium and its subsequent cell division. The phenotype results from elevated expression of HCA2, which encodes a nuclear-localized DNA binding with one finger (Dof) transcription factor Dof5.6. Dof5.6/HCA2 is preferentially expressed in the vasculature of all the organs, particularly in the cambium, phloem, and interfascicular parenchyma cells of inflorescence stems. Dominant-negative analysis further demonstrated that both ubiquitous and in situ repression of HCA2 activity led to disruption of interfascicular cambium formation and development in inflorescence stems. In-depth anatomical analysis showed that HCA2 promotes interfascicular cambium formation at a very early stage of inflorescence stem development. This report demonstrates that a transcription factor gene, HCA2, is involved in regulation of interfascicular cambium formation and vascular tissue development in Arabidopsis.
Figures



Similar articles
-
Transcription factor NTL9 negatively regulates Arabidopsis vascular cambium development during stem secondary growth.Plant Physiol. 2022 Oct 27;190(3):1731-1746. doi: 10.1093/plphys/kiac368. Plant Physiol. 2022. PMID: 35951755 Free PMC article.
-
Cellular events during interfascicular cambium ontogenesis in inflorescence stems of Arabidopsis.Protoplasma. 2014 Sep;251(5):1125-39. doi: 10.1007/s00709-014-0620-5. Epub 2014 Feb 14. Protoplasma. 2014. PMID: 24526327
-
Analysis of secondary growth in the Arabidopsis shoot reveals a positive role of jasmonate signalling in cambium formation.Plant J. 2010 Sep;63(5):811-22. doi: 10.1111/j.1365-313X.2010.04283.x. Plant J. 2010. PMID: 20579310 Free PMC article.
-
From procambium patterning to cambium activation and maintenance in the Arabidopsis root.Curr Opin Plant Biol. 2023 Oct;75:102404. doi: 10.1016/j.pbi.2023.102404. Epub 2023 Jun 21. Curr Opin Plant Biol. 2023. PMID: 37352651 Review.
-
Vascular development in Arabidopsis.Int Rev Cytol. 2002;220:225-56. doi: 10.1016/s0074-7696(02)20007-8. Int Rev Cytol. 2002. PMID: 12224550 Review.
Cited by
-
Identification of SNP Markers Associated with Iron and Zinc Concentrations in Cicer Seeds.Curr Genomics. 2020 Apr;21(3):212-223. doi: 10.2174/1389202921666200413150951. Curr Genomics. 2020. PMID: 33071615 Free PMC article.
-
A systems biology approach to dissect the contribution of brassinosteroid and auxin hormones to vascular patterning in the shoot of Arabidopsis thaliana.Plant Signal Behav. 2010 Jul;5(7):903-6. doi: 10.4161/psb.5.7.12096. Epub 2010 Jul 1. Plant Signal Behav. 2010. PMID: 20622513 Free PMC article.
-
Transcription factor NTL9 negatively regulates Arabidopsis vascular cambium development during stem secondary growth.Plant Physiol. 2022 Oct 27;190(3):1731-1746. doi: 10.1093/plphys/kiac368. Plant Physiol. 2022. PMID: 35951755 Free PMC article.
-
D-myo-inositol-3-phosphate affects phosphatidylinositol-mediated endomembrane function in Arabidopsis and is essential for auxin-regulated embryogenesis.Plant Cell. 2011 Apr;23(4):1352-72. doi: 10.1105/tpc.111.083337. Epub 2011 Apr 19. Plant Cell. 2011. PMID: 21505066 Free PMC article.
-
The family of DOF transcription factors in Brachypodium distachyon: phylogenetic comparison with rice and barley DOFs and expression profiling.BMC Plant Biol. 2012 Nov 5;12:202. doi: 10.1186/1471-2229-12-202. BMC Plant Biol. 2012. PMID: 23126376 Free PMC article.
References
-
- Altamura, M.M., Possenti, M., Matteucci, A., Baima, S., Ruberti, I., and Morelli, G. (2001). Development of the vascular system in the inflorescence stem of Arabidopsis. New Phytol. 151 381–389.
-
- Baima, S., Nobili, F., Sessa, G., Lucchetti, S., Ruberti, I., and Morelli, G. (1995). The expression of the Athb-8 homeobox gene is restricted to provascular cells in Arabidopsis thaliana. Development 121 4171–4182. - PubMed
-
- Baucher, M., El Jaziri, M., and Vandeputte, O. (2007). From primary to secondary growth: Origin and development of the vascular system. J. Exp. Bot. 58 3485–3501. - PubMed
-
- Bonke, M., Thitamadee, S., Mahonen, A.P., Hauser, M.T., and Helariutta, Y. (2003). APL regulates vascular tissue identity in Arabidopsis. Nature 426 181–186. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

