Small-molecule inhibitors target Escherichia coli amyloid biogenesis and biofilm formation
- PMID: 19915538
- PMCID: PMC2838449
- DOI: 10.1038/nchembio.242
Small-molecule inhibitors target Escherichia coli amyloid biogenesis and biofilm formation
Abstract
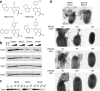
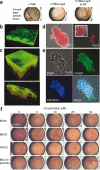
Curli are functional extracellular amyloid fibers produced by uropathogenic Escherichia coli (UPEC) and other Enterobacteriaceae. Ring-fused 2-pyridones, such as FN075 and BibC6, inhibited curli biogenesis in UPEC and prevented the in vitro polymerization of the major curli subunit protein CsgA. The curlicides FN075 and BibC6 share a common chemical lineage with other ring-fused 2-pyridones termed pilicides. Pilicides inhibit the assembly of type 1 pili, which are required for pathogenesis during urinary tract infection. Notably, the curlicides retained pilicide activities and inhibited both curli-dependent and type 1-dependent biofilms. Furthermore, pretreatment of UPEC with FN075 significantly attenuated virulence in a mouse model of urinary tract infection. Curli and type 1 pili exhibited exclusive and independent roles in promoting UPEC biofilms, and curli provided a fitness advantage in vivo. Thus, the ability of FN075 to block the biogenesis of both curli and type 1 pili endows unique anti-biofilm and anti-virulence activities on these compounds.
Figures

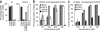
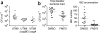
References
Publication types
MeSH terms
Substances
Associated data
Grants and funding
- R37 AI048689/AI/NIAID NIH HHS/United States
- R01 AI048689/AI/NIAID NIH HHS/United States
- K12HD00850/HD/NICHD NIH HHS/United States
- K08DK074443/DK/NIDDK NIH HHS/United States
- R01 AI049950/AI/NIAID NIH HHS/United States
- AI049950/AI/NIAID NIH HHS/United States
- AI073847/AI/NIAID NIH HHS/United States
- P50 DK064540/DK/NIDDK NIH HHS/United States
- K12 HD000850/HD/NICHD NIH HHS/United States
- R01 AI073847/AI/NIAID NIH HHS/United States
- T32A107172/PHS HHS/United States
- R56 AI073847/AI/NIAID NIH HHS/United States
- AI02549/AI/NIAID NIH HHS/United States
- AI048689/AI/NIAID NIH HHS/United States
- P50 DK64540/DK/NIDDK NIH HHS/United States
- K08 DK074443/DK/NIDDK NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources

