Cooperativity of the MUC1 oncoprotein and STAT1 pathway in poor prognosis human breast cancer
- PMID: 19915608
- PMCID: PMC2820589
- DOI: 10.1038/onc.2009.391
Cooperativity of the MUC1 oncoprotein and STAT1 pathway in poor prognosis human breast cancer
Abstract
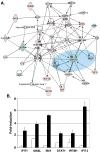
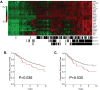
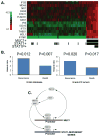
Signal transducer and activator of transcription 1 (STAT1) is activated in the inflammatory response to interferons. The MUC1 oncoprotein is overexpressed in human breast cancers. Analysis of genes differentially expressed in MUC1-transformed cells has identified a network linking MUC1 and STAT1 that is associated with cellular growth and inflammation. The results further show that the MUC1-C subunit associates with STAT1 in cells and the MUC1-C cytoplasmic domain binds directly to the STAT1 DNA-binding domain. The interaction between MUC1-C and STAT1 is inducible by IFNgamma in non-malignant epithelial cells and constitutive in breast cancer cells. Moreover, the MUC1-STAT1 interaction contributes to the activation of STAT1 target genes, including MUC1 itself. Analysis of two independent databases showed that MUC1 and STAT1 are coexpressed in about 15% of primary human breast tumors. Coexpression of MUC1 and the STAT1 pathway was found to be significantly associated with decreased recurrence-free and overall survival. These findings indicate that (i) MUC1 and STAT1 function in an auto-inductive loop, and (ii) activation of both MUC1 and the STAT1 pathway in breast tumors confers a poor prognosis for patients.
Conflict of interest statement
Dr. Kufe has an ownership interest in Genus Oncology and is a consultant to the company.The other authors disclosed no potential conflicts of interest.
Figures


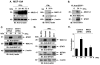

References
-
- Darnell JE, Jr, Kerr IM, Stark GR. Jak-STAT pathways and transcriptional activation in response to IFNs and other extracellular signaling proteins. Science. 1994;264:1415–1421. - PubMed
-
- Deblandre GA, Marinx OP, Evans SS, Majjaj S, Leo O, Caput D, et al. Expression cloning of an interferon-inducible 17-kDa membrane protein implicated in the control of cell growth. 1995;270:23860–23866. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

