Enhanced mRNA cap methylation increases cyclin D1 expression and promotes cell transformation
- PMID: 19915615
- PMCID: PMC3393636
- DOI: 10.1038/onc.2009.368
Enhanced mRNA cap methylation increases cyclin D1 expression and promotes cell transformation
Abstract
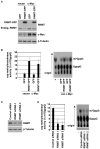
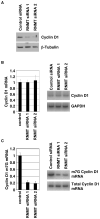
Cap-dependent mRNA translation requires the methylation of the mRNA guanosine cap by RNA guanine-7-methyltransferase (RNMT). mRNA cap methylation was recently described to be rate-limiting for a subset of mRNAs, and to be enhanced by expression of c-Myc and E2F1, although the biological significance of this finding was not investigated. Here, it is reported that increased RNMT expression enhances cellular mRNA cap methyltransferase activity, promotes mammary epithelial cell transformation and cooperates with H-RasV12 or c-Myc to promote fibroblast cell transformation. Cyclin D1 is a prominent oncogene in epithelial tumours. A significant fraction of Cyclin D1 mRNA was found to be unmethylated on the mRNA cap and thus dormant in mammary epithelial cells. Cyclin D1 expression was increased by enhanced mRNA cap methylation. In summary, this report shows that mRNA cap methylation is rate-limiting for expression of an oncogene and cell transformation.
Figures



References
-
- Bentley DL. Rules of engagement: co-transcriptional recruitment of pre-mRNA processing factors. Curr Opin Cell Biol. 2005;17:251–6. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

