Augmentation of reverse transcription by integrase through an interaction with host factor, SIP1/Gemin2 Is critical for HIV-1 infection
- PMID: 19915660
- PMCID: PMC2771899
- DOI: 10.1371/journal.pone.0007825
Augmentation of reverse transcription by integrase through an interaction with host factor, SIP1/Gemin2 Is critical for HIV-1 infection
Abstract
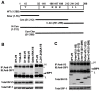
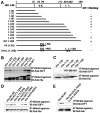
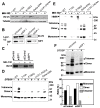
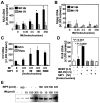
There has been accumulating evidence for the involvement of retroviral integrase (IN) in the reverse transcription of viral RNA. We previously identified a host factor, survival motor neuron-interacting protein 1 (SIP1/Gemin2) that binds to human immunodeficiency virus type 1 (HIV-1) IN and supports HIV-1 infection apparently at reverse transcription step. Here, we demonstrated that HIV-1 IN together with SIP1 augments reverse transcriptase (RT) activity by enhancing the assembly of RT on viral RNA in vitro. Synthetic peptides corresponding to the binding motifs within IN that inhibited the IN-SIP1 interaction abrogated reverse transcription in vitro and in vivo. Furthermore, knockdown of SIP1 reduced intracellular stability and multimer formation of IN through proteasome-mediated degradation machinery. Taken together, SIP1 appears to stabilize functional multimer forms of IN, thereby promoting the assembly of IN and RT on viral RNA to allow efficient reverse transcription, which is a prerequisite for efficient HIV-1 infection.
Conflict of interest statement
Figures
References
-
- Cullen BR. Journey to the center of the cell. Cell. 2001;105:697–700. - PubMed
-
- Goff SP. Intracellular trafficking of retroviral genomes during the early phase of infection: viral exploitation of cellular pathways. J Gene Med. 2001;3:517–528. - PubMed
-
- Katz RA, Skalka AM. The retroviral enzymes. Annu Rev Biochem. 1994;63:133–173. - PubMed
-
- Kalpana GV, Marmon S, Wang W, Crabtree GR, Goff SP. Binding and stimulation of HIV-1 integrase by a human homolog of yeast transcription factor SNF5. Science. 1994;266:2002–2006. - PubMed
-
- Cherepanov P, Maertens G, Proost P, Devreese B, Van Beeumen J, et al. HIV-1 integrase forms stable tetramers and associates with LEDGF/p75 protein in human cells. J Biol Chem. 2003;278:372–381. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials

