Epigenetic signatures associated with different levels of differentiation potential in human stem cells
- PMID: 19915669
- PMCID: PMC2771914
- DOI: 10.1371/journal.pone.0007809
Epigenetic signatures associated with different levels of differentiation potential in human stem cells
Abstract
Background: The therapeutic use of multipotent stem cells depends on their differentiation potential, which has been shown to be variable for different populations. These differences are likely to be the result of key changes in their epigenetic profiles.
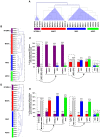
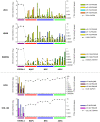
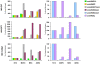
Methodology/principal findings: to address this issue, we have investigated the levels of epigenetic regulation in well characterized populations of pluripotent embryonic stem cells (ESC) and multipotent adult stem cells (ASC) at the trancriptome, methylome, histone modification and microRNA levels. Differences in gene expression profiles allowed classification of stem cells into three separate populations including ESC, multipotent adult progenitor cells (MAPC) and mesenchymal stromal cells (MSC). The analysis of the PcG repressive marks, histone modifications and gene promoter methylation of differentiation and pluripotency genes demonstrated that stem cell populations with a wider differentiation potential (ESC and MAPC) showed stronger representation of epigenetic repressive marks in differentiation genes and that this epigenetic signature was progressively lost with restriction of stem cell potential. Our analysis of microRNA established specific microRNA signatures suggesting specific microRNAs involved in regulation of pluripotent and differentiation genes.
Conclusions/significance: Our study leads us to propose a model where the level of epigenetic regulation, as a combination of DNA methylation and histone modification marks, at differentiation genes defines degrees of differentiation potential from progenitor and multipotent stem cells to pluripotent stem cells.
Conflict of interest statement
Figures




References
-
- Hoffman LM, Carpenter MK. Characterization and culture of human embryonic stem cells. Nat Biotechnol. 2005;23:699–708. - PubMed
-
- Bibikova M, Laurent LC, Ren B, Loring JF, Fan JB. Unraveling epigenetic regulation in embryonic stem cells. Cell Stem Cell. 2008;2:123–134. - PubMed
-
- Bernstein BE, Mikkelsen TS, Xie X, Kamal M, Huebert DJ, et al. A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell. 2006;125:315–326. - PubMed
-
- Pan G, Tian S, Nie J, Yang C, Ruotti V, et al. Whole-genome analysis of histone H3 lysine 4 and lysine 27 methylation in human embryonic stem cells. Cell Stem Cell. 2007;1:299–312. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

