Glioblastoma subclasses can be defined by activity among signal transduction pathways and associated genomic alterations
- PMID: 19915670
- PMCID: PMC2771920
- DOI: 10.1371/journal.pone.0007752
Glioblastoma subclasses can be defined by activity among signal transduction pathways and associated genomic alterations
Abstract
Background: Glioblastoma multiforme (GBM) is an umbrella designation that includes a heterogeneous group of primary brain tumors. Several classification strategies of GBM have been reported, some by clinical course and others by resemblance to cell types either in the adult or during development. From a practical and therapeutic standpoint, classifying GBMs by signal transduction pathway activation and by mutation in pathway member genes may be particularly valuable for the development of targeted therapies.
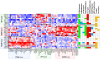
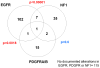
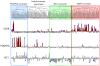
Methodology/principal findings: We performed targeted proteomic analysis of 27 surgical glioma samples to identify patterns of coordinate activation among glioma-relevant signal transduction pathways, then compared these results with integrated analysis of genomic and expression data of 243 GBM samples from The Cancer Genome Atlas (TCGA). In the pattern of signaling, three subclasses of GBM emerge which appear to be associated with predominance of EGFR activation, PDGFR activation, or loss of the RAS regulator NF1. The EGFR signaling class has prominent Notch pathway activation measured by elevated expression of Notch ligands, cleaved Notch receptor, and downstream target Hes1. The PDGF class showed high levels of PDGFB ligand and phosphorylation of PDGFRbeta and NFKB. NF1-loss was associated with lower overall MAPK and PI3K activation and relative overexpression of the mesenchymal marker YKL40. These three signaling classes appear to correspond with distinct transcriptomal subclasses of primary GBM samples from TCGA for which copy number aberration and mutation of EGFR, PDGFRA, and NF1 are signature events.
Conclusions/significance: Proteomic analysis of GBM samples revealed three patterns of expression and activation of proteins in glioma-relevant signaling pathways. These three classes are comprised of roughly equal numbers showing either EGFR activation associated with amplification and mutation of the receptor, PDGF-pathway activation that is primarily ligand-driven, or loss of NF1 expression. The associated signaling activities correlating with these sentinel alterations provide insight into glioma biology and therapeutic strategies.
Conflict of interest statement
Figures

References
-
- Furnari FB, Fenton T, Bachoo RM, Mukasa A, Stommel JM, et al. Malignant astrocytic glioma: genetics, biology, and paths to treatment. Genes Dev. 2007;21:2683–2710. - PubMed
-
- Wen PY, Kesari S. Malignant gliomas in adults. N Engl J Med. 2008;359:492–507. - PubMed
-
- Maher EA, Furnari FB, Bachoo RM, Rowitch DH, Louis DN, et al. Malignant glioma: genetics and biology of a grave matter. Genes Dev. 2001;15:1311–1333. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

