Filamins regulate cell spreading and initiation of cell migration
- PMID: 19915675
- PMCID: PMC2773003
- DOI: 10.1371/journal.pone.0007830
Filamins regulate cell spreading and initiation of cell migration
Abstract
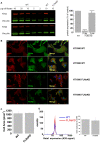
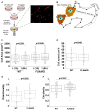
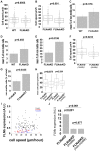
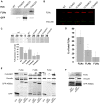
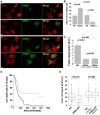
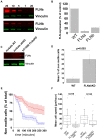
Mammalian filamins (FLNs) are a family of three large actin-binding proteins. FLNa, the founding member of the family, was implicated in migration by cell biological analyses and the identification of FLNA mutations in the neuronal migration disorder periventricular heterotopia. However, recent knockout studies have questioned the relevance of FLNa to cell migration. Here we have used shRNA-mediated knockdown of FLNa, FLNb or FLNa and FLNb, or, alternatively, acute proteasomal degradation of all three FLNs, to generate FLN-deficient cells and assess their ability to migrate. We report that loss of FLNa or FLNb has little effect on migration but that knockdown of FLNa and FLNb, or proteolysis of all three FLNs, impairs migration. The observed defect is primarily a deficiency in initiation of motility rather than a problem with maintenance of locomotion speed. FLN-deficient cells are also impaired in spreading. Re-expression of full length FLNa, but not re-expression of a mutated FLNa lacking immunoglobulin domains 19 to 21, reverts both the spreading and the inhibition of initiation of migration.Our results establish a role for FLNs in cell migration and spreading and suggest that compensation by other FLNs may mask phenotypes in single knockout or knockdown cells. We propose that interactions between FLNs and transmembrane or signalling proteins, mediated at least in part by immunoglobulin domains 19 to 21 are important for both cell spreading and initiation of migration.
Conflict of interest statement
Figures

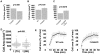

References
-
- Vicente-Manzanares M, Webb DJ, Horwitz AR. Cell migration at a glance. J Cell Sci. 2005;118 - PubMed
-
- Ridley AJ, Schwartz MA, Burridge K, Firtel RA, Ginsberg MH, et al. Cell migration: integrating signals from front to back. Science. 2003;302:1704–1709. - PubMed
-
- Pollard TD, Borisy GG. Cellular motility driven by assembly and disassembly of actin filaments. Cell. 2003;112:453–465. - PubMed
-
- Stossel TP, Condeelis J, Cooley L, Hartwig JH, Noegel A, et al. Filamins as integrators of cell mechanics and signalling. Nat Rev Mol Cell Biol. 2001;2:138–145. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

