Gene regulation and epigenetic remodeling in murine embryonic stem cells by c-Myc
- PMID: 19915707
- PMCID: PMC2773118
- DOI: 10.1371/journal.pone.0007839
Gene regulation and epigenetic remodeling in murine embryonic stem cells by c-Myc
Abstract
Background: The Myc oncoprotein, a transcriptional regulator involved in the etiology of many different tumor types, has been demonstrated to play an important role in the functions of embryonic stem (ES) cells. Nonetheless, it is still unclear as to whether Myc has unique target and functions in ES cells.
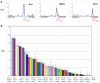
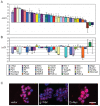
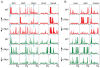
Methodology/principal findings: To elucidate the role of c-Myc in murine ES cells, we mapped its genomic binding sites by chromatin-immunoprecipitation combined with DNA microarrays (ChIP-chip). In addition to previously identified targets we identified genes involved in pluripotency, early development, and chromatin modification/structure that are bound and regulated by c-Myc in murine ES cells. Myc also binds and regulates loci previously identified as Polycomb (PcG) targets, including genes that contain bivalent chromatin domains. To determine whether c-Myc influences the epigenetic state of Myc-bound genes, we assessed the patterns of trimethylation of histone H3-K4 and H3-K27 in mES cells containing normal, increased, and reduced levels of c-Myc. Our analysis reveals widespread and surprisingly diverse changes in repressive and activating histone methylation marks both proximal and distal to Myc binding sites. Furthermore, analysis of bulk chromatin from phenotypically normal c-myc null E7 embryos demonstrates a 70-80% decrease in H3-K4me3, with little change in H3-K27me3, compared to wild-type embryos indicating that Myc is required to maintain normal levels of histone methylation.
Conclusions/significance: We show that Myc induces widespread and diverse changes in histone methylation in ES cells. We postulate that these changes are indirect effects of Myc mediated by its regulation of target genes involved in chromatin remodeling. We further show that a subset of PcG-bound genes with bivalent histone methylation patterns are bound and regulated in response to altered c-Myc levels. Our data indicate that in mES cells c-Myc binds, regulates, and influences the histone modification patterns of genes involved in chromatin remodeling, pluripotency, and differentiation.
Conflict of interest statement
Figures



References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

