Interactive exploration of neuroanatomical meta-spaces
- PMID: 19915734
- PMCID: PMC2776489
- DOI: 10.3389/neuro.11.038.2009
Interactive exploration of neuroanatomical meta-spaces
Abstract
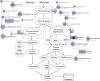
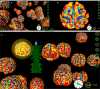
Large-archives of neuroimaging data present many opportunities for re-analysis and mining that can lead to new findings of use in basic research or in the characterization of clinical syndromes. However, interaction with such archives tends to be driven textually, based on subject or image volume meta-data, not the actual neuroanatomical morphology itself, for which the imaging was performed to measure. What is needed is a content-driven approach for examining not only the image content itself but to explore brains that are anatomically similar, and identifying patterns embedded within entire sets of neuroimaging data. With the aim of visual navigation of large- scale neurodatabases, we introduce the concept of brain meta-spaces. The meta-space encodes pair-wise dissimilarities between all individuals in a population and shows the relationships between brains as a navigable framework for exploration. We employ multidimensional scaling (MDS) to implement meta-space processing for a new coordinate system that distributes all data points (brain surfaces) in a common frame-of-reference, with anatomically similar brain data located near each other. To navigate within this derived meta-space, we have developed a fully interactive 3D visualization environment that allows users to examine hundreds of brains simultaneously, visualize clusters of brains with similar characteristics, zoom in on particular instances, and examine the surface topology of an individual brain's surface in detail. The visualization environment not only displays the dissimilarities between brains, but also renders complete surface representations of individual brain structures, allowing an instant 3D view of the anatomies, as well as their differences. The data processing is implemented in a grid-based setting using the LONI Pipeline workflow environment. Additionally users can specify a range of baseline brain atlas spaces as the underlying scale for comparative analyses. The novelty in our approach lies in the user ability to simultaneously view and interact with many brains at once but doing so in a vast meta-space that encodes (dis) similarity in morphometry. We believe that the concept of brain meta-spaces has important implications for the future of how users interact with large-scale archives of primary neuroimaging data.
Keywords: 3D visualization; meta-analysis; neuroanatomical data mining; visual data mining.
Figures



References
-
- Callahan M., Cole M. J., Shepherd J. F., Stinstra J. G., Johnson C. R. (2009). A meshing pipeline for biomedical computing. Eng. Comput. 1, 115–130 10.1007/s00366-008-0106-1 - DOI
-
- Chen J., Zheng T., Thorne W., Zaiane O. R., Goebel R. (2007). Visual data mining of web navigational data. In IV ’07: Proceedings of the 11th International Conference Information Visualization, Washington, DC, IEEE Computer Society, pp. 649–656
-
- Chupin M., Chetelat G., Lemieux L., Dubois B., Garnero L., Benali H., Eustache F., Lehericy S., Desgranges B., Colliot O. (2008). Fully automatic hippocampus segmentation discriminates between early Alzheimer's disease and normal aging. In th IEEE International Symposium on Biomedical Imaging: From Nano to Macro, 2008. ISBI 2008., pp. 97–100
-
- Dinov I., Van Horn J. D., Lozev K. M., Magsipoc R., Petrosyan P., Liu Z., MacKenzie-Graham A., Eggert P., Parker D. S., Toga A. W. (2009). Efficient, distributed and interactive neuroimaging data analysis using the LONI pipeline. Front. Neuroinform. 3, 10.3389/neuro.11.022.2009. - DOI - PMC - PubMed
-
- Dubois B., Feldman H. H., Jacova C., Dekosky S. T., Barberger-Gateau P., Cummings J., Delacourte A., Galasko D., Gauthier S., Jicha G., Meguro K., O'brien J., Pasquier F., Robert P., Rossor M., Salloway S., Stern Y., Visser P. J., Scheltens P. (2007). Research criteria for the diagnosis of Alzheimer's disease: revising the NINCDS-ADRDA criteria. Lancet Neurol. 8, 734–746 10.1016/S1474-4422(07)70178-3 - DOI - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

