Homing endonucleases: from basics to therapeutic applications
- PMID: 19915993
- PMCID: PMC11115532
- DOI: 10.1007/s00018-009-0188-y
Homing endonucleases: from basics to therapeutic applications
Abstract
Homing endonucleases (HE) are double-stranded DNAses that target large recognition sites (12-40 bp). HE-encoding sequences are usually embedded in either introns or inteins. Their recognition sites are extremely rare, with none or only a few of these sites present in a mammalian-sized genome. However, these enzymes, unlike standard restriction endonucleases, tolerate some sequence degeneracy within their recognition sequence. Several members of this enzyme family have been used as templates to engineer tools to cleave DNA sequences that differ from their original wild-type targets. These custom HEs can be used to stimulate double-strand break homologous recombination in cells, to induce the repair of defective genes with very low toxicity levels. The use of tailored HEs opens up new possibilities for gene therapy in patients with monogenic diseases that can be treated ex vivo. This review provides an overview of recent advances in this field.
Figures


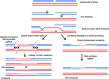
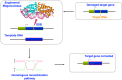


References
-
- Dujon B. Homing nucleases and the yeast mitochondrial omega locus: a historical perspective. Berlin: Springer; 2005.
-
- Liu Q, Belle A, Shub DA, Belfort M, Edgell DR. SegG endonuclease promotes marker exclusion and mediates co-conversion from a distant cleavage site. J Mol Biol. 2003;334:13–23. - PubMed
-
- Edgell DR. Free-standing homing endonucleases of T-even phage: freeloaders or functionaries? Berlin: Springer; 2005.
-
- Edgell DR. Selfish DNA: new abode for homing endonucleases. Curr Biol. 2002;12:R276–R278. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

