A genome-wide deletion mutant screen identifies pathways affected by nickel sulfate in Saccharomyces cerevisiae
- PMID: 19917080
- PMCID: PMC2784802
- DOI: 10.1186/1471-2164-10-524
A genome-wide deletion mutant screen identifies pathways affected by nickel sulfate in Saccharomyces cerevisiae
Abstract
Background: The understanding of the biological function, regulation, and cellular interactions of the yeast genome and proteome, along with the high conservation in gene function found between yeast genes and their human homologues, has allowed for Saccharomyces cerevisiae to be used as a model organism to deduce biological processes in human cells. Here, we have completed a systematic screen of the entire set of 4,733 haploid S. cerevisiae gene deletion strains (the entire set of nonessential genes for this organism) to identify gene products that modulate cellular toxicity to nickel sulfate (NiSO(4)).
Results: We have identified 149 genes whose gene deletion causes sensitivity to NiSO(4) and 119 genes whose gene deletion confers resistance. Pathways analysis with proteins whose absence renders cells sensitive and resistant to nickel identified a wide range of cellular processes engaged in the toxicity of S. cerevisiae to NiSO(4). Functional categories overrepresented with proteins whose absence renders cells sensitive to NiSO(4) include homeostasis of protons, cation transport, transport ATPases, endocytosis, siderophore-iron transport, homeostasis of metal ions, and the diphthamide biosynthesis pathway. Functional categories overrepresented with proteins whose absence renders cells resistant to nickel include functioning and transport of the vacuole and lysosome, protein targeting, sorting, and translocation, intra-Golgi transport, regulation of C-compound and carbohydrate metabolism, transcriptional repression, and chromosome segregation/division. Interactome analysis mapped seven nickel toxicity modulating and ten nickel-resistance networks. Additionally, we studied the degree of sensitivity or resistance of the 111 nickel-sensitive and 72 -resistant strains whose gene deletion product has a similar protein in human cells.
Conclusion: We have undertaken a whole genome approach in order to further understand the mechanism(s) regulating the cell's toxicity to nickel compounds. We have used computational methods to integrate the data and generate global models of the yeast's cellular response to NiSO(4). The results of our study shed light on molecular pathways associated with the cellular response of eukaryotic cells to nickel compounds and provide potential implications for further understanding the toxic effects of nickel compounds to human cells.
Figures
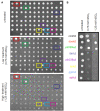

References
-
- Begley TJ, Rosenbach AS, Ideker T, Samson LD. Damage recovery pathways in Saccharomyces cerevisiae revealed by genomic phenotyping and interactome mapping. Mol Cancer Res. 2002;1(2):103–112. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

