The conserved Lysine69 residue plays a catalytic role in Mycobacterium tuberculosis shikimate dehydrogenase
- PMID: 19917104
- PMCID: PMC2789096
- DOI: 10.1186/1756-0500-2-227
The conserved Lysine69 residue plays a catalytic role in Mycobacterium tuberculosis shikimate dehydrogenase
Abstract
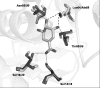
Background: The shikimate pathway is an attractive target for the development of antitubercular agents because it is essential in Mycobacterium tuberculosis, the causative agent of tuberculosis, but absent in humans. M. tuberculosis aroE-encoded shikimate dehydrogenase catalyzes the forth reaction in the shikimate pathway. Structural and functional studies indicate that Lysine69 may be involved in catalysis and/or substrate binding in M. tuberculosis shikimate dehydrogenase. Investigation of the kinetic properties of mutant enzymes can bring important insights about the role of amino acid residues for M. tuberculosis shikimate dehydrogenase.
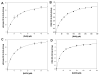
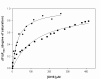
Findings: We have performed site-directed mutagenesis, steady-state kinetics, equilibrium binding measurements and molecular modeling for both the wild-type M. tuberculosis shikimate dehydrogenase and the K69A mutant enzymes. The apparent steady-state kinetic parameters for the M. tuberculosis shikimate dehydrogenase were determined; the catalytic constant value for the wild-type enzyme (50 s-1) is 68-fold larger than that for the mutant K69A (0.73 s-1). There was a modest increase in the Michaelis-Menten constant for DHS (K69A = 76 microM; wild-type = 29 microM) and NADPH (K69A = 30 microM; wild-type = 11 microM). The equilibrium dissociation constants for wild-type and K69A mutant enzymes are 32 (+/- 4) microM and 134 (+/- 21), respectively.
Conclusion: Our results show that the residue Lysine69 plays a catalytic role and is not involved in substrate binding for the M. tuberculosis shikimate dehydrogenase. These efforts on M. tuberculosis shikimate dehydrogenase catalytic mechanism determination should help the rational design of specific inhibitors, aiming at the development of antitubercular drugs.
Figures
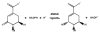
Similar articles
-
Expression, purification and properties of shikimate dehydrogenase from Mycobacterium tuberculosis.J Biochem Mol Biol. 2005 Sep 30;38(5):624-31. doi: 10.5483/bmbrep.2005.38.5.624. J Biochem Mol Biol. 2005. PMID: 16202245
-
Functional shikimate dehydrogenase from Mycobacterium tuberculosis H37Rv: purification and characterization.Protein Expr Purif. 2006 Apr;46(2):429-37. doi: 10.1016/j.pep.2005.10.004. Epub 2005 Oct 27. Protein Expr Purif. 2006. PMID: 16298142
-
Exploring the influence of conserved lysine69 on the catalytic activity of the helicobacter pylori shikimate dehydrogenase: A combined QM/MM and MD simulations.Comput Biol Chem. 2019 Dec;83:107098. doi: 10.1016/j.compbiolchem.2019.107098. Epub 2019 Aug 9. Comput Biol Chem. 2019. PMID: 31421413
-
Shikimate kinase: a potential target for development of novel antitubercular agents.Curr Drug Targets. 2007 Mar;8(3):459-68. doi: 10.2174/138945007780059013. Curr Drug Targets. 2007. PMID: 17348838 Review.
-
Shikimate kinase, a protein target for drug design.Curr Med Chem. 2014;21(5):592-604. doi: 10.2174/09298673113206660299. Curr Med Chem. 2014. PMID: 24164195 Review.
Cited by
-
Mycobacterium tuberculosis Shikimate Pathway Enzymes as Targets for the Rational Design of Anti-Tuberculosis Drugs.Molecules. 2020 Mar 11;25(6):1259. doi: 10.3390/molecules25061259. Molecules. 2020. PMID: 32168746 Free PMC article. Review.
-
Application of modified Michaelis - Menten equations for determination of enzyme inducing and inhibiting drugs.BMC Pharmacol Toxicol. 2021 Oct 11;22(1):57. doi: 10.1186/s40360-021-00521-x. BMC Pharmacol Toxicol. 2021. PMID: 34635182 Free PMC article.
References
-
- Basso LA, Blanchard JS. Resistance to antitubercular drugs. Adv Exp Med Biol. 1998;456:115–144. - PubMed
LinkOut - more resources
Full Text Sources

