A self-consistent description of the conformational behavior of chemically denatured proteins from NMR and small angle scattering
- PMID: 19917239
- PMCID: PMC2776250
- DOI: 10.1016/j.bpj.2009.08.044
A self-consistent description of the conformational behavior of chemically denatured proteins from NMR and small angle scattering
Abstract
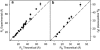
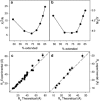
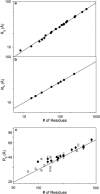
Characterization of the conformational properties of unfolded proteins is essential for understanding the mechanisms of protein folding and misfolding. This information is also fundamental to determining the relationship between flexibility and function in the highly diverse families of intrinsically disordered proteins. Here we present a self-consistent model of conformational sampling of chemically denatured proteins in agreement with experimental data reporting on long-range distance distributions in unfolded proteins using small-angle x-ray scattering and nuclear magnetic resonance pulse-field gradient-based measurements. We find that standard statistical coil models, selected from folded protein databases with secondary structural elements removed, need to be refined to correct backbone dihedral angle sampling of denatured proteins, although they appear to be appropriate for intrinsically disordered proteins. For denatured proteins, pervasive increases in the sampling of more-extended regions of Ramachandran space {50 degrees <psi < 180 degrees} throughout the peptide chain are found to be consistent with all experimental data. These observations are in agreement with previous conclusions derived from short-range nuclear magnetic resonance data from residual dipolar couplings, leading the way to a self-consistent description of denatured chains that is in agreement with short- and long-range data measured using both spectroscopic and scattering techniques.
Figures

References
-
- Tompa P. Intrinsically unstructured proteins. Trends Biochem. Sci. 2002;27:527–533. - PubMed
-
- Fink A.L. Natively unfolded proteins. Curr. Opin. Struct. Biol. 2005;15:35–41. - PubMed
-
- Dyson H.J., Wright P.E. Intrinsically unstructured proteins and their functions. Chem. Rev. 2004;104:3607–3622. - PubMed
-
- Fuxreiter M., Simon I., Friedrich P., Tompa P. Preformed structural elements feature in partner recognition by intrinsically unstructured proteins. J. Mol. Biol. 2004;338:1015–1026. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources

