Tetherin-driven adaptation of Vpu and Nef function and the evolution of pandemic and nonpandemic HIV-1 strains
- PMID: 19917496
- PMCID: PMC2779047
- DOI: 10.1016/j.chom.2009.10.004
Tetherin-driven adaptation of Vpu and Nef function and the evolution of pandemic and nonpandemic HIV-1 strains
Abstract
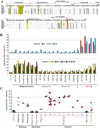
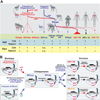
Vpu proteins of pandemic HIV-1 M strains degrade the viral receptor CD4 and antagonize human tetherin to promote viral release and replication. We show that Vpus from SIVgsn, SIVmus, and SIVmon infecting Cercopithecus primate species also degrade CD4 and antagonize tetherin. In contrast, SIVcpz, the immediate precursor of HIV-1, whose Vpu shares a common ancestry with SIVgsn/mus/mon Vpu, uses Nef rather than Vpu to counteract chimpanzee tetherin. Human tetherin, however, is resistant to Nef and thus poses a significant barrier to zoonotic transmission of SIVcpz to humans. Remarkably, Vpus from nonpandemic HIV-1 O strains are poor tetherin antagonists, whereas those from the rare group N viruses do not degrade CD4. Thus, only HIV-1 M evolved a fully functional Vpu following the three independent cross-species transmissions that resulted in HIV-1 groups M, N, and O. This may explain why group M viruses are almost entirely responsible for the global HIV/AIDS pandemic.
Figures




Comment in
-
A tail of Tetherin: how pandemic HIV-1 conquered the world.Cell Host Microbe. 2009 Nov 19;6(5):393-5. doi: 10.1016/j.chom.2009.11.002. Cell Host Microbe. 2009. PMID: 19917491 Free PMC article.
References
-
- Bailes E, Gao F, Bibollet-Ruche F, Courgnaud V, Peeters M, Marx PA, Hahn BH, Sharp PM. Hybrid origin of SIV in chimpanzees. Science. 2003;300:1713. - PubMed
-
- Butler IF, Pandream I, Marxm PA, Apetrei C. HIV genetic diversity: biological and public health consequences. Curr. HIV Res. 2007;5:23–45. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

