Sorting of Drosophila small silencing RNAs partitions microRNA* strands into the RNA interference pathway
- PMID: 19917635
- PMCID: PMC2802036
- DOI: 10.1261/rna.1972910
Sorting of Drosophila small silencing RNAs partitions microRNA* strands into the RNA interference pathway
Abstract
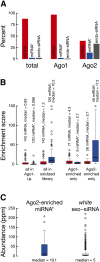
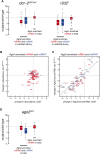
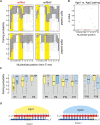
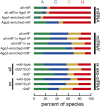
In flies, small silencing RNAs are sorted between Argonaute1 (Ago1), the central protein component of the microRNA (miRNA) pathway, and Argonaute2 (Ago2), which mediates RNA interference. Extensive double-stranded character-as is found in small interfering RNAs (siRNAs)-directs duplexes into Ago2, whereas central mismatches, like those found in miRNA/miRNA* duplexes, direct duplexes into Ago1. Central to this sorting decision is the affinity of the small RNA duplex for the Dcr-2/R2D2 heterodimer, which loads small RNAs into Ago2. Here, we show that while most Drosophila miRNAs are bound to Ago1, miRNA* strands accumulate bound to Ago2. Like siRNA loading, efficient loading of miRNA* strands in Ago2 favors duplexes with a paired central region and requires both Dcr-2 and R2D2. Those miRNA and miRNA* sequences bound to Ago2, like siRNAs diced in vivo from long double-stranded RNA, typically begin with cytidine, whereas Ago1-bound miRNA and miRNA* disproportionately begin with uridine. Consequently, some pre-miRNA generate two or more isoforms from the same side of the stem that differentially partition between Ago1 and Ago2. Our findings provide the first genome-wide test for the idea that Drosophila small RNAs are sorted between Ago1 and Ago2 according to their duplex structure and the identity of their first nucleotide.
Figures



Similar articles
-
R2D2 organizes small regulatory RNA pathways in Drosophila.Mol Cell Biol. 2011 Feb;31(4):884-96. doi: 10.1128/MCB.01141-10. Epub 2010 Dec 6. Mol Cell Biol. 2011. PMID: 21135122 Free PMC article.
-
Sorting of Drosophila small silencing RNAs.Cell. 2007 Jul 27;130(2):299-308. doi: 10.1016/j.cell.2007.05.057. Cell. 2007. PMID: 17662944 Free PMC article.
-
Target RNA-directed tailing and trimming purifies the sorting of endo-siRNAs between the two Drosophila Argonaute proteins.RNA. 2011 Jan;17(1):54-63. doi: 10.1261/rna.2498411. Epub 2010 Nov 24. RNA. 2011. PMID: 21106652 Free PMC article.
-
Diverse small non-coding RNAs in RNA interference pathways.Methods Mol Biol. 2011;764:169-82. doi: 10.1007/978-1-61779-188-8_11. Methods Mol Biol. 2011. PMID: 21748640 Review.
-
Small RNA sorting: matchmaking for Argonautes.Nat Rev Genet. 2011 Jan;12(1):19-31. doi: 10.1038/nrg2916. Epub 2010 Nov 30. Nat Rev Genet. 2011. PMID: 21116305 Free PMC article. Review.
Cited by
-
What Do We Know about the Role of miRNAs in Pediatric Sarcoma?Int J Mol Sci. 2015 Jul 22;16(7):16593-621. doi: 10.3390/ijms160716593. Int J Mol Sci. 2015. PMID: 26204834 Free PMC article. Review.
-
MicroRNA-30e* promotes human glioma cell invasiveness in an orthotopic xenotransplantation model by disrupting the NF-κB/IκBα negative feedback loop.J Clin Invest. 2012 Jan;122(1):33-47. doi: 10.1172/JCI58849. Epub 2011 Dec 12. J Clin Invest. 2012. PMID: 22156201 Free PMC article.
-
Argonaute proteins: structures and their endonuclease activity.Mol Biol Rep. 2021 May;48(5):4837-4849. doi: 10.1007/s11033-021-06476-w. Epub 2021 Jun 11. Mol Biol Rep. 2021. PMID: 34117606 Review.
-
Characterization of rice black-streaked dwarf virus- and rice stripe virus-derived siRNAs in singly and doubly infected insect vector Laodelphax striatellus.PLoS One. 2013 Jun 11;8(6):e66007. doi: 10.1371/journal.pone.0066007. Print 2013. PLoS One. 2013. PMID: 23776591 Free PMC article.
-
Epigenetics and psychostimulant addiction.Cold Spring Harb Perspect Med. 2013 Mar 1;3(3):a012047. doi: 10.1101/cshperspect.a012047. Cold Spring Harb Perspect Med. 2013. PMID: 23359110 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
