Analysis of acyclic nucleoside modifications in siRNAs finds sensitivity at position 1 that is restored by 5'-terminal phosphorylation both in vitro and in vivo
- PMID: 19917641
- PMCID: PMC2811019
- DOI: 10.1093/nar/gkp913
Analysis of acyclic nucleoside modifications in siRNAs finds sensitivity at position 1 that is restored by 5'-terminal phosphorylation both in vitro and in vivo
Abstract
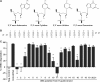
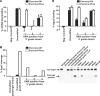
Small interfering RNAs (siRNAs) are short, double-stranded RNAs that use the endogenous RNAi pathway to mediate gene silencing. Phosphorylation facilitates loading of a siRNA into the Ago2 complex and subsequent cleavage of the target mRNA. In this study, 2', 3' seco nucleoside modifications, which contain an acylic ribose ring and are commonly called unlocked nucleic acids (UNAs), were evaluated at all positions along the guide strand of a siRNA targeting apolipoprotein B (ApoB). UNA modifications at positions 1, 2 and 3 were detrimental to siRNA activity. UNAs at positions 1 and 2 prevented phosphorylation by Clp1 kinase, abrogated binding to Ago2, and impaired Ago2-mediated cleavage of the mRNA target. The addition of a 5'-terminal phosphate to siRNA containing a position 1 UNA restored ApoB mRNA silencing, Ago2 binding, and Ago2 mediated cleavage activity. Position 1 UNA modified siRNA containing a 5'-terminal phosphate exhibited a partial restoration of siRNA silencing activity in vivo. These data reveal the complexity of interpreting the effects of chemical modification on siRNA activity, and exemplify the importance of using multiple biochemical, cell-based and in vivo assays to rationally design chemically modified siRNA destined for therapeutic use.
Figures
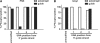

References
-
- Dorsett Y, Tuschl T. siRNAs: applications in functional genomics and potential as therapeutics. Nat. Rev. Drug Discov. 2004;3:318–329. - PubMed
-
- Harborth J, Elbashir SM, Bechert K, Tuschl T, Weber K. Identification of essential genes in cultured mammalian cells using small interfering RNAs. J. Cell Sci. 2001;114:4557–4565. - PubMed
-
- Elbashir SM, Harborth J, Lendeckel W, Yalcin A, Weber K, Tuschl T. Duplexes of 21-nucleotide RNAs mediate RNA interference in cultured mammalian cells. Nature. 2001;411:494–498. - PubMed
-
- Corey DR. RNA learns from antisense. Nat. Chem. Biol. 2007;3:8–11. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

