Mouse Piwi interactome identifies binding mechanism of Tdrkh Tudor domain to arginine methylated Miwi
- PMID: 19918066
- PMCID: PMC2787181
- DOI: 10.1073/pnas.0911640106
Mouse Piwi interactome identifies binding mechanism of Tdrkh Tudor domain to arginine methylated Miwi
Abstract
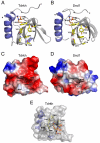
Tudor domains are protein modules that mediate protein-protein interactions, potentially by binding to methylated ligands. A group of germline specific single and multiTudor domain containing proteins (TDRDs) represented by drosophila Tudor and its mammalian orthologs Tdrd1, Tdrd4/RNF17, and Tdrd6 play evolutionarily conserved roles in germinal granule/nuage formation and germ cell specification and differentiation. However, their physiological ligands, and the biochemical and structural basis for ligand recognition, are largely unclear. Here, by immunoprecipitation of endogenous murine Piwi proteins (Miwi and Mili) and proteomic analysis of complexes related to the piRNA pathway, we show that the TDRD group of Tudor proteins are physiological binding partners of Piwi family proteins. In addition, mass spectrometry indicates that arginine residues in RG repeats at the N-termini of Miwi and Mili are methylated in vivo. Notably, we found that Tdrkh/Tdrd2, a novel single Tudor domain containing protein identified in the Miwi complex, is expressed in the cytoplasm of male germ cells and directly associates with Miwi. Mutagenesis studies mapped the Miwi-Tdrkh interaction to the very N-terminal RG/RA repeats of Miwi and showed that the Tdrkh Tudor domain is critical for binding. Furthermore, we have solved the crystal structure of the Tdrkh Tudor domain, which revealed an aromatic binding pocket and negatively charged binding surface appropriate for accommodating methylated arginine. Our findings identify a methylation-directed protein interaction mechanism in germ cells mediated by germline Tudor domains and methylated Piwi family proteins, and suggest a complex mode of regulating the organization and function of Piwi proteins in piRNA silencing pathways.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

References
-
- Maurer-Stroh S, et al. The Tudor domain “royal family”: Tudor, plant agenet, chromo, PWWP and MBT domains. Trends Biochem Sci. 2003;28:69–74. - PubMed
-
- Adams-Cioaba MA, Min J. Structure and function of histone methylation binding proteins. Biochem Cell Biol. 2009;87:93–105. - PubMed
-
- Cote J, Richard S. Tudor domains bind symmetrical dimethylated arginines. J Biol Chem. 2005;280:28476–28483. - PubMed
-
- Huang Y, Fang J, Bedford MT, Zhang Y, Xu RM. Recognition of histone H3 lysine-4 methylation by the double tudor domain of JMJD2A. Science. 2006;312:748–751. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

