A 5' cytosine binding pocket in Puf3p specifies regulation of mitochondrial mRNAs
- PMID: 19918084
- PMCID: PMC2787145
- DOI: 10.1073/pnas.0812079106
A 5' cytosine binding pocket in Puf3p specifies regulation of mitochondrial mRNAs
Abstract
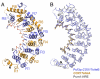
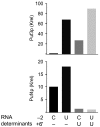
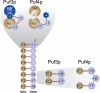
A single regulatory protein can control the fate of many mRNAs with related functions. The Puf3 protein of Saccharomyces cerevisiae is exemplary, as it binds and regulates more than 100 mRNAs that encode proteins with mitochondrial function. Here we elucidate the structural basis of that specificity. To do so, we explore the crystal structures of Puf3p complexes with 2 cognate RNAs. The key determinant of Puf3p specificity is an unusual interaction between a distinctive pocket of the protein with an RNA base outside the "core" PUF-binding site. That interaction dramatically affects binding affinity in vitro and is required for regulation in vivo. The Puf3p structures, combined with those of Puf4p in the same organism, illuminate the structural basis of natural PUF-RNA networks. Yeast Puf3p binds its own RNAs because they possess a -2C and is excluded from those of Puf4p which contain an additional nucleotide in the core-binding site.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


Similar articles
-
Puf3p, a Pumilio family RNA binding protein, localizes to mitochondria and regulates mitochondrial biogenesis and motility in budding yeast.J Cell Biol. 2007 Jan 15;176(2):197-207. doi: 10.1083/jcb.200606054. Epub 2007 Jan 8. J Cell Biol. 2007. PMID: 17210948 Free PMC article.
-
Divergence of Pumilio/fem-3 mRNA binding factor (PUF) protein specificity through variations in an RNA-binding pocket.J Biol Chem. 2012 Feb 24;287(9):6949-57. doi: 10.1074/jbc.M111.326264. Epub 2011 Dec 28. J Biol Chem. 2012. PMID: 22205700 Free PMC article.
-
Architecture and dynamics of overlapped RNA regulatory networks.RNA. 2017 Nov;23(11):1636-1647. doi: 10.1261/rna.062687.117. Epub 2017 Aug 2. RNA. 2017. PMID: 28768715 Free PMC article.
-
Recruitment of the Puf3 protein to its mRNA target for regulation of mRNA decay in yeast.RNA. 2004 Oct;10(10):1625-36. doi: 10.1261/rna.7270204. Epub 2004 Aug 30. RNA. 2004. PMID: 15337848 Free PMC article.
-
The PUF Protein Family: Overview on PUF RNA Targets, Biological Functions, and Post Transcriptional Regulation.Int J Mol Sci. 2018 Jan 30;19(2):410. doi: 10.3390/ijms19020410. Int J Mol Sci. 2018. PMID: 29385744 Free PMC article. Review.
Cited by
-
RNA regulatory networks diversified through curvature of the PUF protein scaffold.Nat Commun. 2015 Sep 14;6:8213. doi: 10.1038/ncomms9213. Nat Commun. 2015. PMID: 26364903 Free PMC article.
-
Integrated analysis of RNA-binding protein complexes using in vitro selection and high-throughput sequencing and sequence specificity landscapes (SEQRS).Methods. 2017 Apr 15;118-119:171-181. doi: 10.1016/j.ymeth.2016.10.001. Epub 2016 Oct 8. Methods. 2017. PMID: 27729296 Free PMC article.
-
Identification of a conserved interface between PUF and CPEB proteins.J Biol Chem. 2012 May 25;287(22):18854-62. doi: 10.1074/jbc.M112.352815. Epub 2012 Apr 11. J Biol Chem. 2012. PMID: 22496444 Free PMC article.
-
Glucose-Regulated Phosphorylation of the PUF Protein Puf3 Regulates the Translational Fate of Its Bound mRNAs and Association with RNA Granules.Cell Rep. 2015 Jun 16;11(10):1638-50. doi: 10.1016/j.celrep.2015.05.014. Epub 2015 Jun 4. Cell Rep. 2015. PMID: 26051939 Free PMC article.
-
Multiple Puf proteins regulate the stability of ribosome biogenesis transcripts.RNA Biol. 2018;15(9):1228-1243. doi: 10.1080/15476286.2018.1521211. Epub 2018 Sep 25. RNA Biol. 2018. PMID: 30251908 Free PMC article.
References
-
- Kimble J, Crittenden SL. Controls of germline stem cells, entry into meiosis, and the sperm/oocyte decision in Caenorhabditis elegans. Annu Rev Cell Dev Biol. 2007;23:405–433. - PubMed
-
- Thompson B, Wickens M, Kimble J. In: Translational Control in Biology and Medicine. Mathews MB, Sonenberg N, Hershey JWB, editors. Cold Spring Harbor, NY: Cold Spring Harbor Lab Press; 2007. pp. 507–544.
-
- Mendez R, Richter JD. Translational control by CPEB: A means to the end. Nat Rev Mol Cell Biol. 2001;2:521–529. - PubMed
-
- Pique M, Lopez JM, Foissac S, Guigo R, Mendez R. A combinatorial code for CPE-mediated translational control. Cell. 2008;132:434–448. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

