Phosphoinositide and inositol phosphate analysis in lymphocyte activation
- PMID: 19918943
- PMCID: PMC4500525
- DOI: 10.1002/0471142735.im1101s87
Phosphoinositide and inositol phosphate analysis in lymphocyte activation
Abstract
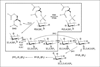
Lymphocyte antigen receptor engagement profoundly changes the cellular content of phosphoinositide lipids and soluble inositol phosphates. Among these, the phosphoinositides phosphatidylinositol 4,5-bisphosphate (PIP2) and phosphatidylinositol 3,4,5-trisphosphate (PIP3) play key signaling roles by acting as pleckstrin homology (PH) domain ligands that recruit signaling proteins to the plasma membrane. Moreover, PIP2 acts as a precursor for the second messenger molecules diacylglycerol and soluble inositol 1,4,5-trisphosphate (IP3), essential mediators of PKC, Ras/Erk, and Ca2+ signaling in lymphocytes. IP3 phosphorylation by IP3 3-kinases generates inositol 1,3,4,5-tetrakisphosphate (IP4), an essential soluble regulator of PH domain binding to PIP3 in developing T cells. Besides PIP2, PIP3, IP3, and IP4, lymphocytes produce multiple other phosphoinositides and soluble inositol phosphates that could have important physiological functions. To aid their analysis, detailed protocols that allow one to simultaneously measure the levels of multiple different phosphoinositide or inositol phosphate isomers in lymphocytes are provided here. They are based on thin layer, conventional and high-performance liquid chromatographic separation methods followed by radiolabeling or non-radioactive metal-dye detection. Finally, less broadly applicable non-chromatographic methods for detection of specific phosphoinositide or inositol phosphate isomers are discussed. Support protocols describe how to obtain pure unstimulated CD4+CD8+ thymocyte populations for analyses of inositol phosphate turnover during positive and negative selection, key steps in T cell development.
Copyright (c) 2009 by John Wiley & Sons, Inc.
Figures





References
-
- Adelt S, Plettenburg O, Stricker R, Reiser G, Altenbach HJ, Vogel G. Enzyme-assisted total synthesis of the optical antipodes d-myo-inositol 3,4,5-trisphosphate and d-myo-inositol 1,5,6-trisphosphate: Aspects of their structure-activity relationship to biologically active inositol phosphates. J. Med. Chem. 1999;42:1262–1273. - PubMed
-
- Alcazar-Roman AR, Wente SR. Inositol polyphosphates: A new frontier for regulating gene expression. Chromosoma. 2008;117:1–13. - PubMed
-
- Amaro R, Escalona A, Murillo M. HPLC with inductively coupled plasma optical emission spectrometric detection for the analysis of inositol phosphates. J. Chromatogr. Sci. 2004;42:491–494. - PubMed
-
- Andrews WV, Conn PM. Measurement of inositol phospholipid metabolites by one-dimensional thin-layer chromatography. Methods Enzymol. 1987;141:156–168. - PubMed
-
- Astoul E, Edmunds C, Cantrell DA, Ward SG. PI 3-K and T-cell activation: Limitations of T-leukemic cell lines as signaling models. Trends Immunol. 2001;22:490–496. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

