Urea destabilizes RNA by forming stacking interactions and multiple hydrogen bonds with nucleic acid bases
- PMID: 19919063
- PMCID: PMC2791195
- DOI: 10.1021/ja905795v
Urea destabilizes RNA by forming stacking interactions and multiple hydrogen bonds with nucleic acid bases
Abstract
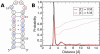
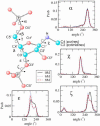
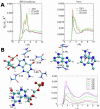
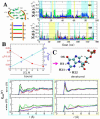
Urea titration of RNA by urea is an effective approach to investigate the forces stabilizing this biologically important molecule. We used all atom molecular dynamics simulations using two urea force fields and two RNA constructs to elucidate in atomic detail the destabilization mechanism of folded RNA in aqueous urea solutions. Urea denatures RNA by forming multiple hydrogen bonds with the RNA bases and has little influence on the phosphodiester backbone. Most significantly we discovered that urea engages in stacking interactions with the bases. We also estimate, for the first time, the m-value for RNA, which is a measure of the strength of urea-RNA interactions. Our work provides a conceptual understanding of the mechanism by which urea enhances RNA folding rates.
Figures
References
-
- Fersht A. Structure and Mechanism in Protein Science: A Guide to Enzyme Catalysis and Protein Folding. W. H. Freeman Company; 1998.
- Tanford C. J. Am. Chem. Soc. 1964;86:2050–2059.
- Makhatadze GI, Privalov PL. J. Mol. Biol. 1992;226:491–505. - PubMed
- Myers JK, Pace CN, Scholtz JM. Protein Sci. 1995;4:2138–2148. - PMC - PubMed
-
- Pan J, Thirumalai D, Woodson SA. J. Mol. Biol. 1997;273:7–13. - PubMed
- Rook MS, Treiber DK, Williamson JR. Proc. Natl. Acad. Sci. 1998;281:609–620. - PubMed
- Shelton VM, Sosnick TR, Pan T. Biochemistry. 1999;38:16831–16839. - PubMed
- Sclavi B, Woodson SA, Sullivan M, Chance MR, Brenowitz M. J. Mol. Biol. 1997;266:144–159. - PubMed
-
- Robinson DR, Jencks WP. J. Am. Chem. Soc. 1965;87:2462–2469. - PubMed
- Wallqvist A, Covell DG, Thirumalai D. J. Am. Chem. Soc. 1998;120:427–428.
- Vanzi F, Madan B, Sharp K. J. Am. Chem. Soc. 1998;120:10748–10753.
- Tobi D, Elber R, Thirumalai D. Biopolymers. 2003;68:359–369. - PubMed
- Soper AK, Castner EW, Luzar A. Biophys. Chem. 2003;105:649–666. - PubMed
- Bennion BJ, Daggett V. Proc. Natl. Acad. Sci. 2003;100:5142–5147. - PMC - PubMed
- O'Brien EP, Dima RI, Brooks B, Thirumalai D. J. Am. Chem. Soc. 2007;129:7346–7353. - PubMed
- Hua L, Zhou R, Thirumalai D, Berne BJ. Proc. Natl. Acad. Sci. 2008;105:16928–16933. - PMC - PubMed
- England JR, Pande VS, Haran G. J. Chem. Soc. 2008;130:11854–11855. - PMC - PubMed
-
- Mason PE, Neilson GW, Enderby JE, Saboungi M-L, Dempsey CE, MacKerell AD, Brady JW. J. Am. Chem. Soc. 2004;126(11):462–11470. - PubMed
-
- Rudisser S, Tinoco I., Jr. J. Mol. Biol. 2000;295:1211–1223. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

