Differential binding and co-binding pattern of FOXA1 and FOXA3 and their relation to H3K4me3 in HepG2 cells revealed by ChIP-seq
- PMID: 19919681
- PMCID: PMC3091322
- DOI: 10.1186/gb-2009-10-11-r129
Differential binding and co-binding pattern of FOXA1 and FOXA3 and their relation to H3K4me3 in HepG2 cells revealed by ChIP-seq
Abstract
Background: The forkhead box/winged helix family members FOXA1, FOXA2, and FOXA3 are of high importance in development and specification of the hepatic linage and the continued expression of liver-specific genes.
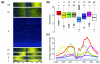
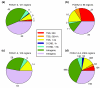
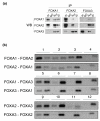
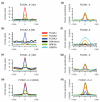
Results: Here, we present a genome-wide location analysis of FOXA1 and FOXA3 binding sites in HepG2 cells through chromatin immunoprecipitation with detection by sequencing (ChIP-seq) studies and compare these with our previous results on FOXA2. We found that these factors often bind close to each other in different combinations and consecutive immunoprecipitation of chromatin for one and then a second factor (ChIP-reChIP) shows that this occurs in the same cell and on the same DNA molecule, suggestive of molecular interactions. Using co-immunoprecipitation, we further show that FOXA2 interacts with both FOXA1 and FOXA3 in vivo, while FOXA1 and FOXA3 do not appear to interact. Additionally, we detected diverse patterns of trimethylation of lysine 4 on histone H3 (H3K4me3) at transcriptional start sites and directionality of this modification at FOXA binding sites. Using the sequence reads at polymorphic positions, we were able to predict allele specific binding for FOXA1, FOXA3, and H3K4me3. Finally, several SNPs associated with diseases and quantitative traits were located in the enriched regions.
Conclusions: We find that ChIP-seq can be used not only to create gene regulatory maps but also to predict molecular interactions and to inform on the mechanisms for common quantitative variation.
Figures


References
-
- Kaestner KH, Knochel W, Martinez DE. Unified nomenclature for the winged helix/forkhead transcription factors. Genes Dev. 2000;14:142–146. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

