Disparate oxidant gene expression of airway epithelium compared to alveolar macrophages in smokers
- PMID: 19919714
- PMCID: PMC2787510
- DOI: 10.1186/1465-9921-10-111
Disparate oxidant gene expression of airway epithelium compared to alveolar macrophages in smokers
Abstract
Background: The small airway epithelium and alveolar macrophages are exposed to oxidants in cigarette smoke leading to epithelial dysfunction and macrophage activation. In this context, we asked: what is the transcriptome of oxidant-related genes in small airway epithelium and alveolar macrophages, and does their response differ substantially to inhaled cigarette smoke?
Methods: Using microarray analysis, with TaqMan RT-PCR confirmation, we assessed oxidant-related gene expression in small airway epithelium and alveolar macrophages from the same healthy nonsmoker and smoker individuals.
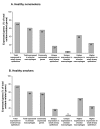
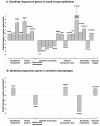
Results: Of 155 genes surveyed, 87 (56%) were expressed in both cell populations in nonsmokers, with higher expression in alveolar macrophages (43%) compared to airway epithelium (24%). In smokers, there were 15 genes (10%) up-regulated and 7 genes (5%) down-regulated in airway epithelium, but only 3 (2%) up-regulated and 2 (1%) down-regulated in alveolar macrophages. Pathway analysis of airway epithelium showed oxidant pathways dominated, but in alveolar macrophages immune pathways dominated.
Conclusion: Thus, the response of different cell-types with an identical genome exposed to the same stress of smoking is different; responses of alveolar macrophages are more subdued than those of airway epithelium. These findings are consistent with the observation that, while the small airway epithelium is vulnerable, alveolar macrophages are not "diseased" in response to smoking.
Trial registration: ClinicalTrials.gov ID: NCT00224185 and NCT00224198.
Figures



References
Publication types
MeSH terms
Associated data
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

