Estrogen-mediated regulation of Igf1 transcription and uterine growth involves direct binding of estrogen receptor alpha to estrogen-responsive elements
- PMID: 19920132
- PMCID: PMC2807324
- DOI: 10.1074/jbc.M109.043471
Estrogen-mediated regulation of Igf1 transcription and uterine growth involves direct binding of estrogen receptor alpha to estrogen-responsive elements
Abstract
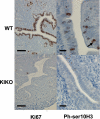
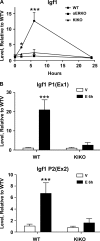
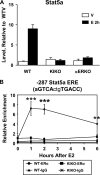
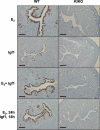
Estrogen enables uterine proliferation, which depends on synthesis of the IGF1 growth factor. This proliferation and IGF1 synthesis requires the estrogen receptor (ER), which binds directly to target DNA sequences (estrogen-responsive elements or EREs), or interacts with other transcription factors, such as AP1, to impact transcription. We observe neither uterine growth nor an increase in Igf1 transcript in a mouse with a DNA-binding mutated ER alpha (KIKO), indicating that both Igf1 regulation and uterine proliferation require the DNA binding function of the ER. We identified several potential EREs in the Igf1 gene, and chromatin immunoprecipitation analysis revealed ER alpha binding to these EREs in wild type but not KIKO chromatin. STAT5 is also reported to regulate Igf1; uterine Stat5a transcript is increased by estradiol (E(2)), but not in KIKO or alpha ERKO uteri, indicating ER alpha- and ERE-dependent regulation. ER alpha binds to a potential Stat5a ERE. We hypothesize that E(2) increases Stat5a transcript through ERE binding; that ER alpha, either alone or together with STAT5, then acts to increase Igf1 transcription; and that the resulting lack of IGF1 impairs KIKO uterine growth. Treatment with exogenous IGF1, alone or in combination with E(2), induces proliferation in wild type but not KIKO uteri, indicating that IGF1 replacement does not rescue the KIKO proliferative response. Together, these observations suggest in contrast to previous in vitro studies of IGF-1 regulation involving AP1 motifs that direct ER alpha-DNA interaction is required to increase Igf1 transcription. Additionally, full ER alpha function is needed to mediate other cellular signals of the growth factor for uterine growth.
Figures




Similar articles
-
Requirement of estrogen receptor-alpha in insulin-like growth factor-1 (IGF-1)-induced uterine responses and in vivo evidence for IGF-1/estrogen receptor cross-talk.J Biol Chem. 2002 Mar 8;277(10):8531-7. doi: 10.1074/jbc.M109592200. Epub 2001 Dec 21. J Biol Chem. 2002. PMID: 11751931
-
A distal super enhancer mediates estrogen-dependent mouse uterine-specific gene transcription of Igf1 (insulin-like growth factor 1).J Biol Chem. 2019 Jun 21;294(25):9746-9759. doi: 10.1074/jbc.RA119.008759. Epub 2019 May 9. J Biol Chem. 2019. PMID: 31073032 Free PMC article.
-
Novel DNA motif binding activity observed in vivo with an estrogen receptor α mutant mouse.Mol Endocrinol. 2014 Jun;28(6):899-911. doi: 10.1210/me.2014-1051. Epub 2014 Apr 8. Mol Endocrinol. 2014. PMID: 24713037 Free PMC article.
-
Selective disruption of ER{alpha} DNA-binding activity alters uterine responsiveness to estradiol.Mol Endocrinol. 2009 Dec;23(12):2111-6. doi: 10.1210/me.2009-0356. Epub 2009 Oct 7. Mol Endocrinol. 2009. PMID: 19812388 Free PMC article.
-
Non-nuclear actions of estrogen: new targets for prevention and treatment of cardiovascular disease.Mol Interv. 2002 Jul;2(4):219-28. doi: 10.1124/mi.2.4.219. Mol Interv. 2002. PMID: 14993393 Free PMC article. Review.
Cited by
-
Sequential activation of uterine epithelial IGF1R by stromal IGF1 and embryonic IGF2 directs normal uterine preparation for embryo implantation.J Mol Cell Biol. 2021 Dec 6;13(9):646-661. doi: 10.1093/jmcb/mjab034. J Mol Cell Biol. 2021. PMID: 34097060 Free PMC article.
-
Assessment of Zearalenone-Induced Cell Survival and of Global Gene Regulation in Mouse TM4 Sertoli Cells.Toxins (Basel). 2022 Jan 26;14(2):98. doi: 10.3390/toxins14020098. Toxins (Basel). 2022. PMID: 35202126 Free PMC article.
-
A tissue-specific role of membrane-initiated ERα signaling for the effects of SERMs.J Endocrinol. 2022 Mar 29;253(2):75-84. doi: 10.1530/JOE-21-0398. J Endocrinol. 2022. PMID: 35256537 Free PMC article.
-
17β-Estradiol and ICI182,780 Differentially Regulate STAT5 Isoforms in Female Mammary Epithelium, With Distinct Outcomes.J Endocr Soc. 2018 Feb 26;2(3):293-309. doi: 10.1210/js.2017-00399. eCollection 2018 Mar 1. J Endocr Soc. 2018. PMID: 29594259 Free PMC article.
-
Altered gene expression patterns during the initiation and promotion stages of neonatally diethylstilbestrol-induced hyperplasia/dysplasia/neoplasia in the hamster uterus.Reprod Toxicol. 2014 Dec;50:68-86. doi: 10.1016/j.reprotox.2014.09.002. Epub 2014 Sep 19. Reprod Toxicol. 2014. PMID: 25242112 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

