EWS/FLI and its downstream target NR0B1 interact directly to modulate transcription and oncogenesis in Ewing's sarcoma
- PMID: 19920188
- PMCID: PMC2789197
- DOI: 10.1158/0008-5472.CAN-09-1540
EWS/FLI and its downstream target NR0B1 interact directly to modulate transcription and oncogenesis in Ewing's sarcoma
Abstract
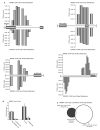
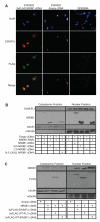
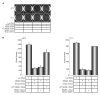
Most Ewing's sarcomas harbor chromosomal translocations that encode fusions between EWS and ETS family members. The most common fusion, EWS/FLI, consists of an EWSR1-derived strong transcriptional activation domain fused, in-frame, to the DNA-binding domain-containing portion of FLI1. EWS/FLI functions as an aberrant transcription factor to regulate genes that mediate the oncogenic phenotype of Ewing's sarcoma. One of these regulated genes, NR0B1, encodes a corepressor protein, and likely plays a transcriptional role in tumorigenesis. However, the genes that NR0B1 regulates and the transcription factors it interacts with in Ewing's sarcoma are largely unknown. We used transcriptional profiling and chromatin immunoprecipitation to identify genes that are regulated by NR0B1, and compared these data to similar data for EWS/FLI. Although the transcriptional profile overlapped as expected, we also found that the genome-wide localization of NR0B1 and EWS/FLI overlapped as well, suggesting that they regulate some genes coordinately. Further analysis revealed that NR0B1 and EWS/FLI physically interact. This protein-protein interaction is likely to be relevant for the development of Ewing's sarcoma because mutations in NR0B1 that disrupt the interaction have transcriptional consequences and also abrogate oncogenic transformation. Taken together, these data suggest that EWS/FLI and NR0B1 physically interact, coordinately modulate gene expression, and mediate the transformed phenotype of Ewing's sarcoma.
Figures



References
-
- Turc-Carel C, Aurias A, Mugneret F, et al. Chromosomes in Ewing's sarcoma. I. An evaluation of 85 cases of remarkable consistency of t(11;22)(q24;q12) Cancer Genet Cytogenet. 1988;32(2):229–38. - PubMed
-
- Kovar H, Aryee DN, Jug G, et al. EWS/FLI-1 antagonists induce growth inhibition of Ewing tumor cells in vitro. Cell Growth Differ. 1996;7(4):429–37. - PubMed
-
- Ouchida M, Ohno T, Fujimura Y, Rao VN, Reddy ES. Loss of tumorigenicity of Ewing's sarcoma cells expressing antisense RNA to EWS-fusion transcripts. Oncogene. 1995;11(6):1049–54. - PubMed
-
- Smith R, Owen LA, Trem DJ, et al. Expression profiling of EWS/FLI identifies NKX2.2 as a critical target gene in Ewing's sarcoma. Cancer Cell. 2006;9(5):405–16. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

