Positive cross-talk between estrogen receptor and NF-kappaB in breast cancer
- PMID: 19920189
- PMCID: PMC2996265
- DOI: 10.1158/0008-5472.CAN-09-2608
Positive cross-talk between estrogen receptor and NF-kappaB in breast cancer
Erratum in
- Cancer Res. 2010 Jan 15;70(2):854
- Cancer Res. 2010 Mar 1;70(5):2140
Abstract
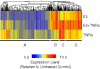
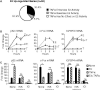
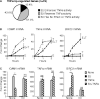
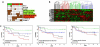
Estrogen receptors (ER) and nuclear factor-kappaB (NF-kappaB) are known to play important roles in breast cancer, but these factors are generally thought to repress each other's activity. However, we have recently found that ER and NF-kappaB can also act together in a positive manner to synergistically increase gene transcription. To examine the extent of cross-talk between ER and NF-kappaB, a microarray study was conducted in which MCF-7 breast cancer cells were treated with 17beta-estradiol (E(2)), tumor necrosis factor alpha (TNFalpha), or both. Follow-up studies with an ER antagonist and NF-kappaB inhibitors show that cross-talk between E(2) and TNFalpha is mediated by these two factors. We find that although transrepression between ER and NF-kappaB does occur, positive cross-talk is more prominent with three gene-specific patterns of regulation: (a) TNFalpha enhances E(2) action on approximately 30% of E(2)-upregulated genes; (b) E(2) enhances TNFalpha activity on approximately 15% of TNFalpha-upregulated genes; and (c) E(2) + TNFalpha causes a more than additive upregulation of approximately 60 genes. Consistent with their prosurvival roles, ER and NF-kappaB and their target gene, BIRC3, are involved in protecting breast cancer cells against apoptosis. Furthermore, genes positively regulated by E(2) + TNFalpha are clinically relevant because they are enriched in luminal B breast tumors and their expression profiles can distinguish a cohort of patients with poor outcome following endocrine treatment. Taken together, our findings suggest that positive cross-talk between ER and NF-kappaB is more extensive than anticipated and that these factors may act together to promote survival of breast cancer cells and progression to a more aggressive phenotype.
Figures


Similar articles
-
CBP mediates NF-κB-dependent histone acetylation and estrogen receptor recruitment to an estrogen response element in the BIRC3 promoter.Mol Cell Biol. 2012 Jan;32(2):569-75. doi: 10.1128/MCB.05869-11. Epub 2011 Nov 14. Mol Cell Biol. 2012. PMID: 22083956 Free PMC article.
-
NF-kappaB activation in inflammatory breast cancer is associated with oestrogen receptor downregulation, secondary to EGFR and/or ErbB2 overexpression and MAPK hyperactivation.Br J Cancer. 2007 Sep 3;97(5):659-69. doi: 10.1038/sj.bjc.6603906. Epub 2007 Aug 14. Br J Cancer. 2007. PMID: 17700572 Free PMC article.
-
Similar NF-κB gene signatures in TNF-α treated human endothelial cells and breast tumor biopsies.PLoS One. 2011;6(7):e21589. doi: 10.1371/journal.pone.0021589. Epub 2011 Jul 6. PLoS One. 2011. PMID: 21754991 Free PMC article.
-
Minireview: Inflammation: an instigator of more aggressive estrogen receptor (ER) positive breast cancers.Mol Endocrinol. 2012 Mar;26(3):360-71. doi: 10.1210/me.2011-1302. Epub 2012 Feb 2. Mol Endocrinol. 2012. PMID: 22301780 Free PMC article. Review.
-
Linkage between EGFR family receptors and nuclear factor kappaB (NF-kappaB) signaling in breast cancer.J Cell Physiol. 2006 Dec;209(3):645-52. doi: 10.1002/jcp.20785. J Cell Physiol. 2006. PMID: 17001676 Review.
Cited by
-
Genome-wide progesterone receptor binding: cell type-specific and shared mechanisms in T47D breast cancer cells and primary leiomyoma cells.PLoS One. 2012;7(1):e29021. doi: 10.1371/journal.pone.0029021. Epub 2012 Jan 17. PLoS One. 2012. PMID: 22272226 Free PMC article.
-
Fucoxanthin and Its Metabolite Fucoxanthinol in Cancer Prevention and Treatment.Mar Drugs. 2015 Jul 31;13(8):4784-98. doi: 10.3390/md13084784. Mar Drugs. 2015. PMID: 26264004 Free PMC article. Review.
-
Estrogen Receptor Beta: The Promising Biomarker and Potential Target in Metastases.Int J Mol Sci. 2021 Feb 6;22(4):1656. doi: 10.3390/ijms22041656. Int J Mol Sci. 2021. PMID: 33562134 Free PMC article. Review.
-
Mechanistic analysis of enhancer sequences in the estrogen receptor transcriptional program.Commun Biol. 2024 Jun 11;7(1):719. doi: 10.1038/s42003-024-06400-5. Commun Biol. 2024. PMID: 38862711 Free PMC article.
-
Molecular Pathways: The Balance between Cancer and the Immune System Challenges the Therapeutic Specificity of Targeting Nuclear Factor-κB Signaling for Cancer Treatment.Clin Cancer Res. 2016 Sep 1;22(17):4302-8. doi: 10.1158/1078-0432.CCR-15-1374. Epub 2016 Jul 15. Clin Cancer Res. 2016. PMID: 27422962 Free PMC article. Review.
References
-
- Clarke R, Liu MC, Bouker KB, et al. Antiestrogen resistance in breast cancer and the role of estrogen receptor signaling. Oncogene. 2003;22:7316–39. - PubMed
-
- EBCTCG Effects of chemotherapy and hormonal therapy for early breast cancer on recurrence and 15-year survival: an overview of the randomised trials. Lancet. 2005;365:1687–717. - PubMed
-
- Kuukasjarvi T, Kononen J, Helin H, Holli K, Isola J. Loss of estrogen receptor in recurrent breast cancer is associated with poor response to endocrine therapy. J Clin Oncol. 1996;14:2584–9. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

