Th22 cells represent a distinct human T cell subset involved in epidermal immunity and remodeling
- PMID: 19920355
- PMCID: PMC2786807
- DOI: 10.1172/JCI40202
Th22 cells represent a distinct human T cell subset involved in epidermal immunity and remodeling
Abstract
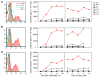
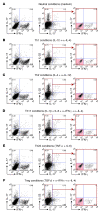
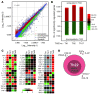
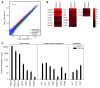
Th subsets are defined according to their production of lineage-indicating cytokines and functions. In this study, we have identified a subset of human Th cells that infiltrates the epidermis in individuals with inflammatory skin disorders and is characterized by the secretion of IL-22 and TNF-alpha, but not IFN-gamma, IL-4, or IL-17. In analogy to the Th17 subset, cells with this cytokine profile have been named the Th22 subset. Th22 clones derived from patients with psoriasis were stable in culture and exhibited a transcriptome profile clearly separate from those of Th1, Th2, and Th17 cells; it included genes encoding proteins involved in tissue remodeling, such as FGFs, and chemokines involved in angiogenesis and fibrosis. Primary human keratinocytes exposed to Th22 supernatants expressed a transcriptome response profile that included genes involved in innate immune pathways and the induction and modulation of adaptive immunity. These proinflammatory Th22 responses were synergistically dependent on IL-22 and TNF-alpha. Furthermore, Th22 supernatants enhanced wound healing in an in vitro injury model, which was exclusively dependent on IL-22. In conclusion, the human Th22 subset may represent a separate T cell subset with a distinct identity with respect to gene expression and function, present within the epidermal layer in inflammatory skin diseases. Future strategies directed against the Th22 subset may be of value in chronic inflammatory skin disorders.
Figures




References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

