Dissection of a complex transcriptional response using genome-wide transcriptional modelling
- PMID: 19920812
- PMCID: PMC2795478
- DOI: 10.1038/msb.2009.84
Dissection of a complex transcriptional response using genome-wide transcriptional modelling
Abstract
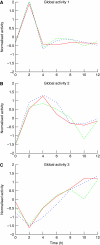
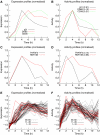
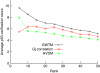
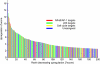
Modern genomics technologies generate huge data sets creating a demand for systems level, experimentally verified, analysis techniques. We examined the transcriptional response to DNA damage in a human T cell line (MOLT4) using microarrays. By measuring both mRNA accumulation and degradation over a short time course, we were able to construct a mechanistic model of the transcriptional response. The model predicted three dominant transcriptional activity profiles-an early response controlled by NFkappaB and c-Jun, a delayed response controlled by p53, and a late response related to cell cycle re-entry. The method also identified, with defined confidence limits, the transcriptional targets associated with each activity. Experimental inhibition of NFkappaB, c-Jun and p53 confirmed that target predictions were accurate. Model predictions directly explained 70% of the 200 most significantly upregulated genes in the DNA-damage response. Genome-wide transcriptional modelling (GWTM) requires no prior knowledge of either transcription factors or their targets. GWTM is an economical and effective method for identifying the main transcriptional activators in a complex response and confidently predicting their targets.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures




References
-
- Abe Y, Takeuchi T, Kagawa-Miki L, Ueda N, Shigemoto K, Yasukawa M, Kito K (2007) A mitotic kinase TOPK enhances Cdk1/cyclin B1-dependent phosphorylation of PRC1 and promotes cytokinesis. J Mol Biol 370: 231–245 - PubMed
-
- Barenco M, Papouli E, Shah S, Brewer D, Miller CJ, Hubank M (2009) rHVDM: an R package to predict the activity and targets of a transcription factor. Bioinformatics 25: 419–420 - PubMed
-
- Bates S, Rowan S, Vousden KH (1996) Characterisation of human cyclin G1 and G2: DNA damage inducible genes. Oncogene 13: 1103–1109 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

