Why proteins without an alpha-crystallin domain should not be included in the human small heat shock protein family HSPB
- PMID: 19921466
- PMCID: PMC3082639
- DOI: 10.1007/s12192-009-0155-4
Why proteins without an alpha-crystallin domain should not be included in the human small heat shock protein family HSPB
Abstract
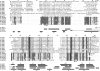
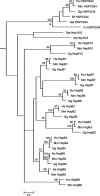
The presence of an alpha-crystallin domain documents the evolutionary relatedness of the ubiquitous family of small heat shock proteins. Sequence and three-dimensional structure provide no evidence for the presence of such a domain in HSPC034, recently proposed as the 11th member of the human HSPB family. Also, phylogenetic analyses detect no relationship between HSPC034 and the human HSPB1-10 sequences. Arguments are provided as to why inclusion in the HSPB family of proteins like HSPC034, which resemble small heat shock proteins in being heat-inducible and having chaperone-like properties and a low monomeric mass, but are evolutionarily unrelated, is misleading and confusing.
Figures
References
-
- Bellyei S, Szigeti A, Boronkai A, Pozsgai E, Gomori E, Melegh B, Janaky T, Bognar Z, Hocsak E, Sumegi B, Gallyas F., Jr Inhibition of cell death by a novel 16.2 kD heat shock protein predominantly via Hsp90 mediated lipid rafts stabilization and Akt activation pathway. Apoptosis. 2007;12:97–112. doi: 10.1007/s10495-006-0486-x. - DOI - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

