Repeatability and reproducibility in proteomic identifications by liquid chromatography-tandem mass spectrometry
- PMID: 19921851
- PMCID: PMC2818771
- DOI: 10.1021/pr9006365
Repeatability and reproducibility in proteomic identifications by liquid chromatography-tandem mass spectrometry
Abstract
The complexity of proteomic instrumentation for LC-MS/MS introduces many possible sources of variability. Data-dependent sampling of peptides constitutes a stochastic element at the heart of discovery proteomics. Although this variation impacts the identification of peptides, proteomic identifications are far from completely random. In this study, we analyzed interlaboratory data sets from the NCI Clinical Proteomic Technology Assessment for Cancer to examine repeatability and reproducibility in peptide and protein identifications. Included data spanned 144 LC-MS/MS experiments on four Thermo LTQ and four Orbitrap instruments. Samples included yeast lysate, the NCI-20 defined dynamic range protein mix, and the Sigma UPS 1 defined equimolar protein mix. Some of our findings reinforced conventional wisdom, such as repeatability and reproducibility being higher for proteins than for peptides. Most lessons from the data, however, were more subtle. Orbitraps proved capable of higher repeatability and reproducibility, but aberrant performance occasionally erased these gains. Even the simplest protein digestions yielded more peptide ions than LC-MS/MS could identify during a single experiment. We observed that peptide lists from pairs of technical replicates overlapped by 35-60%, giving a range for peptide-level repeatability in these experiments. Sample complexity did not appear to affect peptide identification repeatability, even as numbers of identified spectra changed by an order of magnitude. Statistical analysis of protein spectral counts revealed greater stability across technical replicates for Orbitraps, making them superior to LTQ instruments for biomarker candidate discovery. The most repeatable peptides were those corresponding to conventional tryptic cleavage sites, those that produced intense MS signals, and those that resulted from proteins generating many distinct peptides. Reproducibility among different instruments of the same type lagged behind repeatability of technical replicates on a single instrument by several percent. These findings reinforce the importance of evaluating repeatability as a fundamental characteristic of analytical technologies.
Figures



 LTQ@73
LTQ@73  LTQx@65
LTQx@65  LTQ2@95
LTQ2@95  LTQc@65
LTQc@65  Orbi@86
Orbi@86  OrbiO@65
OrbiO@65  OrbiP@65
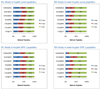
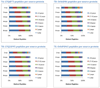
OrbiP@65  OrbiW@56 The number of spectra matched to peptide sequences varied considerably from instrument to instrument. These four graphs show the identification success for each yeast replicate in each instrument for Studies 5, 6, and 8 (two concentrations). LTQs are colored blue, and Orbitraps are shown in pink. A different shape represents each instrument appearing in the studies, as described in the legend. Each symbol reports identifications from an individual technical replicate. Despite SOP controls in Studies 5 and 6, instruments differed from each other by large margins. The Orbitrap at site 86 delivered the highest performance in Study 5, but performance decreased using the higher flow rate specified by the SOP in Study 6. Increasing the yeast concentration by five-fold in Study 8 increased the numbers of spectra identified.
OrbiW@56 The number of spectra matched to peptide sequences varied considerably from instrument to instrument. These four graphs show the identification success for each yeast replicate in each instrument for Studies 5, 6, and 8 (two concentrations). LTQs are colored blue, and Orbitraps are shown in pink. A different shape represents each instrument appearing in the studies, as described in the legend. Each symbol reports identifications from an individual technical replicate. Despite SOP controls in Studies 5 and 6, instruments differed from each other by large margins. The Orbitrap at site 86 delivered the highest performance in Study 5, but performance decreased using the higher flow rate specified by the SOP in Study 6. Increasing the yeast concentration by five-fold in Study 8 increased the numbers of spectra identified.






References
-
- Steen H, Mann M. The ABC's (and XYZ's) of peptide sequencing. Nat Rev Mol Cell Biol. 2004;5(9):699–711. - PubMed
-
- Prakash A, Mallick P, Whiteaker J, Zhang H, Paulovich A, Flory M, Lee H, Aebersold R, Schwikowski B. Signal maps for mass spectrometry-based comparative proteomics. Mol Cell Proteomics. 2006;5(3):423–432. - PubMed
-
- Liu H, Sadygov RG, Yates JR., 3rd A model for random sampling and estimation of relative protein abundance in shotgun proteomics. Anal Chem. 2004;76(14):4193–4201. - PubMed
-
- Tabb DL, MacCoss MJ, Wu CC, Anderson SD, Yates JR., 3rd Similarity among tandem mass spectra from proteomic experiments: detection, significance, and utility. Anal Chem. 2003;75(10):2470–2477. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

