Phylogeographic reconstruction of a bacterial species with high levels of lateral gene transfer
- PMID: 19922616
- PMCID: PMC2784454
- DOI: 10.1186/1741-7007-7-78
Phylogeographic reconstruction of a bacterial species with high levels of lateral gene transfer
Abstract
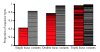
Background: Phylogeographic reconstruction of some bacterial populations is hindered by low diversity coupled with high levels of lateral gene transfer. A comparison of recombination levels and diversity at seven housekeeping genes for eleven bacterial species, most of which are commonly cited as having high levels of lateral gene transfer shows that the relative contributions of homologous recombination versus mutation for Burkholderia pseudomallei is over two times higher than for Streptococcus pneumoniae and is thus the highest value yet reported in bacteria. Despite the potential for homologous recombination to increase diversity, B. pseudomallei exhibits a relative lack of diversity at these loci. In these situations, whole genome genotyping of orthologous shared single nucleotide polymorphism loci, discovered using next generation sequencing technologies, can provide very large data sets capable of estimating core phylogenetic relationships. We compared and searched 43 whole genome sequences of B. pseudomallei and its closest relatives for single nucleotide polymorphisms in orthologous shared regions to use in phylogenetic reconstruction.
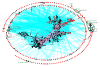
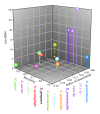
Results: Bayesian phylogenetic analyses of >14,000 single nucleotide polymorphisms yielded completely resolved trees for these 43 strains with high levels of statistical support. These results enable a better understanding of a separate analysis of population differentiation among >1,700 B. pseudomallei isolates as defined by sequence data from seven housekeeping genes. We analyzed this larger data set for population structure and allele sharing that can be attributed to lateral gene transfer. Our results suggest that despite an almost panmictic population, we can detect two distinct populations of B. pseudomallei that conform to biogeographic patterns found in many plant and animal species. That is, separation along Wallace's Line, a biogeographic boundary between Southeast Asia and Australia.
Conclusion: We describe an Australian origin for B. pseudomallei, characterized by a single introduction event into Southeast Asia during a recent glacial period, and variable levels of lateral gene transfer within populations. These patterns provide insights into mechanisms of genetic diversification in B. pseudomallei and its closest relatives, and provide a framework for integrating the traditionally separate fields of population genetics and phylogenetics for other bacterial species with high levels of lateral gene transfer.
Figures




References
-
- Alland D, Whittam TS, Murray MB, Cave MD, Hazbon MH, Dix K, Kokoris M, Duesterhoeft A, Eisen JA, Fraser CM, Fleischmann RD. Modeling bacterial evolution with comparative-genome-based marker systems: application to Mycobacterium tuberculosis evolution and pathogenesis. J Bacteriol. 2003;185:3392–3399. doi: 10.1128/JB.185.11.3392-3399.2003. - DOI - PMC - PubMed
-
- Kennedy AD, Otto M, Braughton KR, Whitney AR, Chen L, Mathema B, Mediavilla JR, Byrne KA, Parkins LD, Tenover FC. Epidemic community-associated methicillin-resistant Staphylococcus aureus: recent clonal expansion and diversification. Proc Natl Acad Sci USA. 2008;105:1327–1332. doi: 10.1073/pnas.0710217105. - DOI - PMC - PubMed
-
- Pearson T, Busch JD, Ravel J, Read TD, Rhoton SD, U'Ren JM, Simonson TS, Kachur SM, Leadem RR, Cardon ML. Phylogenetic discovery bias in Bacillus anthracis using single-nucleotide polymorphisms from whole-genome sequencing. Proc Natl Acad Sci USA. 2004;101:13536–13541. doi: 10.1073/pnas.0403844101. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

