Multimerization of hepatitis delta antigen is a critical determinant of RNA binding specificity
- PMID: 19923178
- PMCID: PMC2812348
- DOI: 10.1128/JVI.01723-09
Multimerization of hepatitis delta antigen is a critical determinant of RNA binding specificity
Abstract
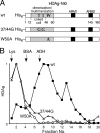
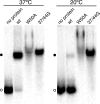
Hepatitis delta virus (HDV) RNA forms an unbranched rod structure that is associated with hepatitis delta antigen (HDAg) in cells replicating HDV. Previous in vitro binding experiments using bacterially expressed HDAg showed that the formation of a minimal ribonucleoprotein complex requires an HDV unbranched rod RNA of at least about 300 nucleotides (nt) and suggested that HDAg binds the RNA as a multimer of fixed size. The present study specifically examines the role of HDAg multimerization in the formation of the HDV ribonucleoprotein complex (RNP). Disruption of HDAg multimerization by site-directed mutagenesis was found to profoundly alter the nature of RNP formation. Mutant HDAg proteins defective for multimerization exhibited neither the 300-nt RNA size requirement for binding nor specificity for the unbranched rod structure. The results unambiguously demonstrate that HDAg binds HDV RNA as a multimer and that the HDAg multimer is formed prior to binding the RNA. RNP formation was found to be temperature dependent, which is consistent with conformational changes occurring on binding. Finally, analysis of RNPs constructed with unbranched rod RNAs successively longer than the minimum length indicated that multimeric binding is not limited to the first HDAg bound and that a minimum RNA length of between 604 and 714 nt is required for binding of a second multimer. The results confirm the previous proposal that HDAg binds as a large multimer and demonstrate that the multimer is a critical determinant of the structure of the HDV RNP.
Figures




References
-
- Antson, A. A., E. J. Dodson, G. Dodson, R. B. Greaves, X. Chen, and P. Gollnick. 1999. Structure of the trp RNA-binding attenuation protein, TRAP, bound to RNA. Nature 401:235-242. - PubMed
-
- Baumann, C., J. Otridge, and P. Gollnick. 1996. Kinetic and thermodynamic analysis of the interaction between TRAP (trp RNA-binding attenuation protein) of Bacillus subtilis and trp leader RNA. J. Biol. Chem. 271:12269-12274. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

