Comparing models of evolution for ordered and disordered proteins
- PMID: 19923193
- PMCID: PMC2822292
- DOI: 10.1093/molbev/msp277
Comparing models of evolution for ordered and disordered proteins
Erratum in
- Mol Biol Evol. 2012 Jan;29(1):443
Abstract
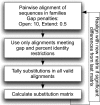
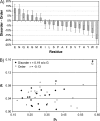
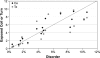
Most models of protein evolution are based upon proteins that form relatively rigid 3D structures. A significant fraction of proteins, the so-called disordered proteins, do not form rigid 3D structures and sample a broad conformational ensemble. Disordered proteins do not typically maintain long-range interactions, so the constraints on their evolution should be different than ordered proteins. To test this hypothesis, we developed and compared models of evolution for disordered and ordered proteins. Substitution matrices were constructed using the sequences of putative homologs for sets of experimentally characterized disordered and ordered proteins. Separate matrices, at three levels of sequence similarity (>85%, 85-60%, and 60-40%), were inferred for each type of protein structure. The substitution matrices for disordered and ordered proteins differed significantly at each level of sequence similarity. The disordered matrices reflected a greater likelihood of evolutionary changes, relative to the ordered matrices, and these changes involved nonconservative substitutions. Glutamic acid and asparagine were interesting exceptions to this result. Important differences between the substitutions that are accepted in disordered proteins relative to ordered proteins were also identified. In general, disordered proteins have fewer evolutionary constraints than ordered proteins. However, some residues like tryptophan and tyrosine are highly conserved in disordered proteins. This is due to their important role in forming protein-protein interfaces. Finally, the amino acid frequencies for disordered proteins, computed during the development of the matrices, were compared with amino acid frequencies for different categories of secondary structure in ordered proteins. The highest correlations were observed between the amino acid frequencies in disordered proteins and the solvent-exposed loops and turns of ordered proteins, supporting an emerging structural model for disordered proteins.
Figures




Similar articles
-
Markov models of amino acid substitution to study proteins with intrinsically disordered regions.PLoS One. 2011;6(5):e20488. doi: 10.1371/journal.pone.0020488. Epub 2011 May 27. PLoS One. 2011. PMID: 21647374 Free PMC article.
-
Amino acid substitution scoring matrices specific to intrinsically disordered regions in proteins.Sci Rep. 2019 Nov 8;9(1):16380. doi: 10.1038/s41598-019-52532-8. Sci Rep. 2019. PMID: 31704957 Free PMC article.
-
Aligning protein sequence and analysing substitution pattern using a class-specific matrix.J Biosci. 2010 Jun;35(2):295-314. doi: 10.1007/s12038-010-0033-3. J Biosci. 2010. PMID: 20689185
-
Substitution scoring matrices for proteins - An overview.Protein Sci. 2020 Nov;29(11):2150-2163. doi: 10.1002/pro.3954. Epub 2020 Oct 12. Protein Sci. 2020. PMID: 32954566 Free PMC article. Review.
-
Bioinformatical approaches to characterize intrinsically disordered/unstructured proteins.Brief Bioinform. 2010 Mar;11(2):225-43. doi: 10.1093/bib/bbp061. Epub 2009 Dec 10. Brief Bioinform. 2010. PMID: 20007729 Review.
Cited by
-
Polymorphism Analysis Reveals Reduced Negative Selection and Elevated Rate of Insertions and Deletions in Intrinsically Disordered Protein Regions.Genome Biol Evol. 2015 Jun 4;7(6):1815-26. doi: 10.1093/gbe/evv105. Genome Biol Evol. 2015. PMID: 26047845 Free PMC article.
-
Evolutionary patterns in coiled-coils.Genome Biol Evol. 2015 Jan 10;7(2):545-56. doi: 10.1093/gbe/evv007. Genome Biol Evol. 2015. PMID: 25577198 Free PMC article.
-
Intrinsically disordered regions of p53 family are highly diversified in evolution.Biochim Biophys Acta. 2013 Apr;1834(4):725-38. doi: 10.1016/j.bbapap.2013.01.012. Epub 2013 Jan 22. Biochim Biophys Acta. 2013. PMID: 23352836 Free PMC article.
-
The case for intrinsically disordered proteins playing contributory roles in molecular recognition without a stable 3D structure.F1000 Biol Rep. 2013;5:1. doi: 10.3410/B5-1. Epub 2013 Jan 11. F1000 Biol Rep. 2013. PMID: 23361308 Free PMC article.
-
Increased polymorphism near low-complexity sequences across the genomes of Plasmodium falciparum isolates.Genome Biol Evol. 2011;3:539-50. doi: 10.1093/gbe/evr045. Epub 2011 May 21. Genome Biol Evol. 2011. PMID: 21602572 Free PMC article.
References
-
- Anisimova M, Kosiol C. Investigating protein-coding sequence evolution with probabilistic codon substitution models. Mol Biol Evol. 2009;26:255–271. - PubMed
-
- Appella E, Anderson CW. Post-translational modifications and activation of p53 by genotoxic stresses. Eur J Biochem. 2001;268:2764–2772. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

