Structure of a eukaryotic nonribosomal peptide synthetase adenylation domain that activates a large hydroxamate amino acid in siderophore biosynthesis
- PMID: 19923209
- PMCID: PMC2807299
- DOI: 10.1074/jbc.M109.071324
Structure of a eukaryotic nonribosomal peptide synthetase adenylation domain that activates a large hydroxamate amino acid in siderophore biosynthesis
Abstract
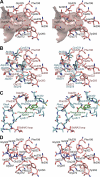
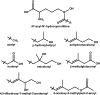
Nonribosomal peptide synthetases (NRPSs) are large, multidomain proteins that are involved in the biosynthesis of an array of secondary metabolites. We report the structure of the third adenylation domain from the siderophore-synthesizing NRPS, SidN, from the endophytic fungus Neotyphodium lolii. This is the first structure of a eukaryotic NRPS domain, and it reveals a large binding pocket required to accommodate the unusual amino acid substrate, N(delta)-cis-anhydromevalonyl-N(delta)-hydroxy-L-ornithine (cis-AMHO). The specific activation of cis-AMHO was confirmed biochemically, and an AMHO moiety was unambiguously identified as a component of the fungal siderophore using mass spectroscopy. The protein structure shows that the substrate binding pocket is defined by 17 amino acid residues, in contrast to both prokaryotic adenylation domains and to previous predictions based on modeling. Existing substrate prediction methods for NRPS adenylation domains fail for domains from eukaryotes due to the divergence of their signature sequences from those of prokaryotes. Thus, this new structure will provide a basis for improving prediction methods for eukaryotic NRPS enzymes that play important and diverse roles in the biology of fungi.
Figures




References
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources

