The structure of the mammalian RNase H2 complex provides insight into RNA.NA hybrid processing to prevent immune dysfunction
- PMID: 19923215
- PMCID: PMC2823502
- DOI: 10.1074/jbc.M109.059048
The structure of the mammalian RNase H2 complex provides insight into RNA.NA hybrid processing to prevent immune dysfunction
Abstract
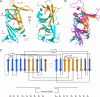
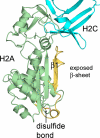
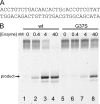
The mammalian RNase H2 ribonuclease complex has a critical function in nucleic acid metabolism to prevent immune activation with likely roles in processing of RNA primers in Okazaki fragments during DNA replication, in removing ribonucleotides misinserted by DNA polymerases, and in eliminating RNA.DNA hybrids during cell death. Mammalian RNase H2 is a heterotrimeric complex of the RNase H2A, RNase H2B, and RNase H2C proteins that are all required for proper function and activity. Mutations in the human RNase H2 genes cause Aicardi-Goutières syndrome. We have determined the crystal structure of the three-protein mouse RNase H2 enzyme complex to better understand the molecular basis of RNase H2 dysfunction in human autoimmunity. The structure reveals the intimately interwoven architecture of RNase H2B and RNase H2C that interface with RNase H2A in a complex ideally suited for nucleic acid binding and hydrolysis coupled to protein-protein interaction motifs that could allow for efficient participation in multiple cellular functions. We have identified four conserved acidic residues in the active site that are necessary for activity and suggest a two-metal ion mechanism of catalysis for RNase H2. An Okazaki fragment has been modeled into the RNase H2 nucleic acid binding site providing insight into the recognition of RNA.DNA junctions by the RNase H2. Further structural and biochemical analyses show that some RNase H2 disease-causing mutations likely result in aberrant protein-protein interactions while the RNase H2A subunit-G37S mutation appears to distort the active site accounting for the demonstrated substrate specificity modification.
Figures
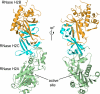


Similar articles
-
Aicardi-Goutières syndrome: clues from the RNase H2 knock-out mouse.J Mol Med (Berl). 2013 Nov;91(11):1235-40. doi: 10.1007/s00109-013-1061-x. Epub 2013 Jun 7. J Mol Med (Berl). 2013. PMID: 23744109 Review.
-
Functional consequences of the RNase H2A subunit mutations that cause Aicardi-Goutieres syndrome.J Biol Chem. 2011 May 13;286(19):16984-91. doi: 10.1074/jbc.M111.228833. Epub 2011 Mar 16. J Biol Chem. 2011. PMID: 21454563 Free PMC article.
-
The structural and biochemical characterization of human RNase H2 complex reveals the molecular basis for substrate recognition and Aicardi-Goutières syndrome defects.J Biol Chem. 2011 Mar 25;286(12):10540-50. doi: 10.1074/jbc.M110.181974. Epub 2010 Dec 22. J Biol Chem. 2011. PMID: 21177858 Free PMC article.
-
Crystal structure of RNase H3-substrate complex reveals parallel evolution of RNA/DNA hybrid recognition.Nucleic Acids Res. 2014 Aug;42(14):9285-94. doi: 10.1093/nar/gku615. Epub 2014 Jul 12. Nucleic Acids Res. 2014. PMID: 25016521 Free PMC article.
-
Structural Studies of RNases H2 as an Example of Crystal Structure Determination of Protein-Nucleic Acid Complexes.Methods Enzymol. 2017;592:123-143. doi: 10.1016/bs.mie.2017.03.009. Epub 2017 May 8. Methods Enzymol. 2017. PMID: 28668118 Review.
Cited by
-
Identification of host proteins interacting with the integrin-like A domain of Toxoplasma gondii micronemal protein MIC2 by yeast-two-hybrid screening.Parasit Vectors. 2014 Nov 26;7:543. doi: 10.1186/s13071-014-0543-1. Parasit Vectors. 2014. PMID: 25423901 Free PMC article.
-
Aicardi-Goutières syndrome gene Rnaseh2c is a metastasis susceptibility gene in breast cancer.PLoS Genet. 2019 May 24;15(5):e1008020. doi: 10.1371/journal.pgen.1008020. eCollection 2019 May. PLoS Genet. 2019. PMID: 31125342 Free PMC article.
-
Large-Scale Photolithographic Synthesis of Chimeric DNA/RNA Hairpin Microarrays To Explore Sequence Specificity Landscapes of RNase HII Cleavage.Biochemistry. 2019 Nov 5;58(44):4389-4397. doi: 10.1021/acs.biochem.9b00806. Epub 2019 Oct 28. Biochemistry. 2019. PMID: 31631649 Free PMC article.
-
Effects of neutral salts and pH on the activity and stability of human RNase H2.J Biochem. 2017 Sep 1;162(3):211-219. doi: 10.1093/jb/mvx021. J Biochem. 2017. PMID: 28402412 Free PMC article.
-
Aicardi-Goutières syndrome: clues from the RNase H2 knock-out mouse.J Mol Med (Berl). 2013 Nov;91(11):1235-40. doi: 10.1007/s00109-013-1061-x. Epub 2013 Jun 7. J Mol Med (Berl). 2013. PMID: 23744109 Review.
References
-
- Ogawa T., Okazaki T. (1980) Annu. Rev. Biochem. 49, 421–457 - PubMed
-
- Okazaki T., Kurosawa Y., Ogawa T., Seki T., Shinozaki K., Hirose S., Fujiyama A., Kohara Y., Machida Y., Tamanoid F., Hozumi T. (1979) Cold Spring Harb. Symp. Quant. Biol. 43, 203–219 - PubMed
-
- Rossi M. L., Purohit V., Brandt P. D., Bambara R. A. (2006) Chem. Rev. 106, 453–473 - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

